Journal of Animal Health and Production
Research Article
Differences in Melanogenesis-related Gene Expression between Korean Brindle Cattle and Korean Native Cattle for Coat Color Decisions
Kyoung Sub Jung1, Yun Dong Choi2, Sang Hwan Kim3*
1ChungCheongBuk-Do Province Veterinary Service Laboratory, Cheongju, Chungcheongbuk-do, 28153, Korea; 2Seoul Hoseo Occupational Training College, Seoul, 07583, Korea; 3Institute of Genetic Engineering, Hankyong National University, Ansung, Gyeonggi-do, 17579, Korea.
Kyoung Sub Jung and Yun Dong Choi contributed equally to this work.
Abstract | In melanocytes, the expression of different pigments is controlled by the melanocortin 1 receptor (MC1R: ED, E+, and e) and the agouti locus alleles. This study aimed to identify melanogenesis-associated genes that are differentially expressed in the yellow and brindle coat of the Korean native cattle. The expression level of MC1R protein in the Korean brindle cattle (KBC) and the Korean dark cattle (KDC: Black coat color) was evaluated and it was found that the MC1R protein level in KDC was much higher than expected, but the expression was found to be lower in the large sweat glands present in the epidermis. The results of the RT-PCR analysis demonstrated that the mRNA expression level of MC1R in the KBC was completely correlated to the localization analysis. The expression level of MC1R mRNA was found to be elevated in the Korean Hanwoo cattle (KHC) compared to those in other groups. Moreover, the genes involved in the MAPK vs. Wnt signaling pathway are differentially expressed between KBC and KHC. The expression of melanogenesis-associated genes was higher in the KBC, while the Wnt signal pathways and other minor genes were highly expressed in KHC. From these results, we were able to demonstrate the diversity in Korean native cattle by assessing the differences in the expression of melanogenesis-associated genes.
Keywords | Melanogenesis, MARK, Wnt, Coat color, Korean brindle cattle
Editor | Asghar Ali Kamboh, Sindh Agriculture University, Tandojam, Pakistan.
Received | March 23, 2021; Accepted | July 04, 2021; Published | August 25, 2021
*Correspondence | Sang Hwan Kim, Institute of Genetic Engineering, Hankyong National University, Ansung, Gyeonggi-do, 17579, Korea; Email: [email protected]
Citation | Jung KS, Choi YD, Kim SH (2021). Differences in melanogenesis-related gene expression between korean brindle cattle and Korean native cattle for coat color decisions. J. Anim. Health Prod. 9(3): 321-330.
DOI | http://dx.doi.org/10.17582/journal.jahp/2021/9.3.321.330
ISSN | 2308-2801
Copyright © 2021 Jung et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
It has previously been reported that determining the breed of the livestock and securing the genetic resources are very important aspects for the national livestock industry. The most important criteria for determining the species of livestock is the characterization of the coat color that remains unchanged even after generations. Although extensive research has been carried out for characterizing the coat color, that on the coat color of Korean brindle cattle (KBC) is still ongoing. However, several opinions have been shared on the coat color decision of KBC. In particular, out of the 70 genes that were identified to be related to the coat color (Jackson, 1994), three genes are known to regulate each other to control the coat color decision. Based on the results of Kim et al. (2014), it has been demonstrated that the interaction of MAPK and Wnt signaling pathway with the melanogenesis pathway is crucial for determining the coat color. Controlling the genes related to the synthesis of melanin in pigment cells such as tyrosinase (TYR), tyrosinase-released protein 1 (TRP-1), and dopachrome tautomerase (DCT) (Guibert et al., 2004; Wang and Hebert, 2006) could play a crucial role in the synthesis of pheomelanin and eumelanin. In several studies, the most common method used for analyzing coat color is to determine the genetic local patterns of the melanocortin 1 receptor (MC1R) gene. MC1R gene is located on the chromosome 18 of the cow genome and is reported to play a pivotal role in the expression of pheomelanin or eumelanin, thereby affecting pigmentation (Jackson, 1993; Cone et al., 1996; Do et al., 2007). The MC1R gene, which is also known as the Extension locus (E locus), plays a crucial role in the manifestation of coat color by regulating the synthesis of pheomelanin and eumelanin, which is mainly linked to the three antagonistic variations of this gene, including ED, E+, and e (Jackson, 1993; Robbins et al., 1993; Royo et al., 2005). In the case of yellow and black in native Korean cattle, expression of “e”, which is a dominant allele accompanied by missense mutation, leads to the appearance of black color mainly due to the synthesis of eumelanin, whereas expression of E+, a wild type allele, leads to the appearance of various forms of coat color, and expression of e, an allelic variant with homogenous junction caused by a frameshift mutation, leads to the appearance of red coat color (Klungland et al., 1995; Chung et al., 2000; Kim et al., 2000; Jin et al., 2011). However, to our knowledge, no existing reports show that the expression of MC1R within the hair roots varies by breed, and there is a huge lack of understanding regarding the genetic processes that might occur in the hair roots. Therefore, in this study, the genetic pattern and the expression pattern of melanogenesis related genes were analyzed in the hair roots of KBC.
MATERIALS AND METHODS
Animals
The Korean cattle used in this study were all raised at a farm affiliated to the National Institute of Animal Health Korea and were categorized based on the coat color as brindle, black, and yellow according to the classification method described in (Kim et al., 2013) (Figure 1). First, the difference in coat color was analyzed by evaluating the MC1R expression and the variation in the expression pattern of the E locus of MC1R, using a total of 325 heads (cattle with identified relatives (Table 1). The epidermal tissues from brindle cattle (brindle color: 10 heads), dark cattle (black color: 10 heads), Hanwoo (yellow color: 10 heads), and dairy cattle (Holstein: 10 heads) were collected and used for performing the gene expression analyses and histological analysis.
DNA extraction
The blood was collected in a tube containing 5 mg of heparin and was carried to the laboratory in a container with ice. The red blood cells were removed from 4 ml of blood by adding the RBC lysis buffer (10 mM Tris, 10 mM NaCl, 5 mM MgCl2), the white blood cells were transferred to a 1.5 ml tube. DNA was extracted using the Invitrogen Easy DNA Kit (Invitrogen, CA, USA), according to the manufacturer’s instructions. Lastly, 40 μg/ml of RNase was added to the DNA to remove the residual RNA and incubated for 30 min at 37 °C. The extracted DNA was stored at 4 °C until further experimentation.
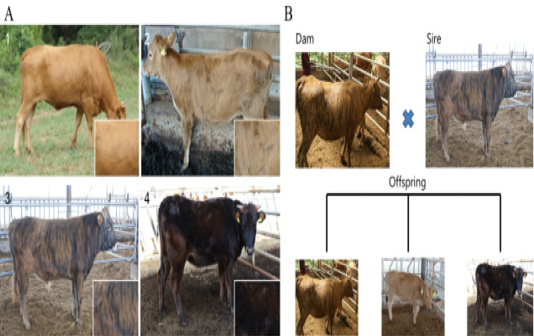
Figure 1: Different patterns of expression of coat color between Korean native cattle (Kim et al., 2013). A: Differences in the cattle coat color determined by the ratio of Yellow: Black. 1) Hanwoo (10:0), 2) Korean brindle cattle (Brindle coat color type 1 (9:1)), 3) Korean brindle cattle (Brindle coat color type 2 (4:6)). B: The difference in the coat color of the offsprings was evaluated based on the crossbreeding of the cattle.
Genotypic analysis of MC1R using PCR-RFLP
The PCR for analyzing MC1R genotype was performed using a PCR thermal cycler (Takara, Shiga, Japan). It was quantified by mixing the genomic DNA concentration of about 300 ± 30 ng/μl, 10 pmol MC1R (Gene Bank accession no: AF445642) forward/reverse primer (forward primer sequence: CAGTGCCTGGAGGTGTCCAT, reverse primer sequence: GGCCAGCATGTGGACGTAGA), 4 μl of 2.5 mM dNTP, 10× buffer, 16.5 μl distilled water, 2 U Taq polymerase (Toyobo, Osaka, Japan), in a total of 25 μl reaction mixture. The reaction conditions were pre-denaturation for 10 min at 95 °C; followed by 35 cycles of denaturation for 30 s at 95 °C, annealing for 30 s at 60 °C, and extension for 1 min at 72 °C; and final extension for 5 min at 72 °C. To analyze the genetic variation of MC1R, 10 μl of the PCR product was mixed with 2 U of MspI (Toyobo, Osaka, Japan) and 2 μl of 10× buffer for 4 h at 37 °C. After the enzyme treatment, the samples were electrophoresed using 2% agarose gel, and the patterns of the DNA band were assessed to classify as E+/E+, E+/e and e/e, and estimated by their genetic variation of MC1R, according to the method described by Klungland et al. (1995) and Park et al. (2012).
Table 1: Differences between Sire and Dam based on the MC1R genotype in Korean brindle cattle.
| GROUP | Coat color | No. of animals | MC1R genotype | |||
| Pattern | Classification code | E+/E+ | E+/e | e/e | ||
| A | Brindle | 1 | 81 | 60 | 21 | |
| 2 | 33 | 16 | 17 | |||
| 3 | 55 | 21 | 29 | |||
| Sub total | 164(50.46%) | 97(62.99%) | 67(48.20%) | 0 | ||
| B | Yellowish brown | 4 | 65 | 12 | 21 | 32 |
| Sub total | 65(20.00%) | 12(7.79%) | 21(15.11%) | 32(100%) | ||
| C | Dark brown | 5 | 30 | 13 | 17 | |
| 6 | 31 | 9 | 22 | |||
| 7 | 35 | 23 | 12 | |||
| Sub total | 96(29.54%) | 45(29.22%) | 51(36.69%) | 0 | ||
| Total | 325 | 154 | 139 | 32 | ||
Real-time polymerase chain reaction
Reverse transcription and PCR amplification were performed using the one-step SYBR RT-PCR Kit (TaKaRa, Shiga, Japan). To perform the RT-PCR, complementary DNA (cDNA) was prepared using the mRNA extracted from the epidermal tissue of the different groups that was collected according to the phenotype of each coat color. cDNA amplification performed using Bos_18sr primer as an internal control for normalization and relative quantitation. The expression of each gene was analyzed using the quantitative values of the cDNA amplification. Primers for amplification of genes of interest were prepared as listed in Table 4. The PCR was performed at an annealing temperature 57~60 °C with 35 cycles. The cycle threshold (Ct) values in the semi-log amplification plot of the geometric region were considered if they did not exceed 30, and the abnormal values were excluded from the analysis. A comparative Ct method was applied to perform the analysis using the method described by Schmittgen and Livak (2008), in which the fold change values were converted and verified.
Protein extraction
Total proteins were extracted using the PRO-PREPTM Kit (Intron biotechnology, Jungwon, KOR), according to the manufacturer’s instructions. Briefly, the samples were homogenized in 400μl of Pro-prep solution and allowed to chill on the ice for 20 min. The insoluble material was removed by centrifugation (600 x g, 5min, 4°C), and the supernatant was transferred to a fresh 1.5ml tube. The total protein content was quantified using the Bradford protein assay (Bio-Rad, CA, USA), and then the final protein samples were stored at -80°C.
Western blot analysis
Total protein of 30 μg from each sample was separated on 13% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) in duplicates, and then the separated proteins were transferred onto an Immunoblot PVDF membrane (Bio-Rad, CA, USA). The membrane was incubated with the blocking buffer (3% Skim milk) for overnight at 4 °C. Later, the membrane was washed using the washing buffer (1× Tris + 1× NaCl + 0.05% Tween 20) one time for 10 min. The membrane was then incubated with the antibodies: MC1R (diluted 1:1000; Host: Rabbit polyclonal, Abcam, Cambridge, UK), MITF (diluted 1:1000; Host: Rabbit polyclonal, Abcam), and β-actin (diluted 1:5000; Host: Rabbit polyclonal, Abcam) for 2 h. After this step, the membranes were washed using 1× TBS-T buffer three times for 10 min each, and then were incubated with HRP-conjugated anti-rabbit secondary antibody (diluted 1:5000; Abcam). Thereafter, the membranes were visualized in a dark room using the ECL detection kit. The membrane was incubating in the detection reagent for 5 min, the reagent was drained off, and the membranes were exposed using a sheet of diagnostic film in the cassette for 1–5 min; protein expression was calculated using the software Alpha Innotech ver. 4.0.0 (San Leandro, CA, USA) and was normalized to that of β-actin, which served as an internal control.
Immunohistochemistry (IHC)
Paraffin sections were dewaxed in a xylene substitute (Polyclear solvent; Polysciences, Warrington, PA) and rehydrated using a gradient of ethanol. Antigen unmasking was performed by heating the samples at 95 °C in 10 mM sodium citrate (pH 6.0). Endogenous peroxidases were quenched using 3% hydrogen peroxide. Sections were incubated (overnight, 4 °C) with the antibodies, MC1R (diluted 1:300; Abcam, MA) and MITF (diluted 1:300; Abcam, MA). Washed sections were then incubated with anti-rabbit secondary antibody (diluted 1:300; Abcam, MA) for 1 h at Room temperature (RT), rinsed and incubated with ABC detection reagent (Vactor, USA) for 10 min. Diaminobenzidene (DAB) was used as a substrate for the enzyme horseradish peroxidase (HRP). Sections were then counter stained with PAW reagent and Harris hematoxylin (Protocol) prepared in 4% acetic acid. Tissues were dehydrated and cleared and then the cover slips were placed in Permount (Fisher, Pittsburgh, PA, USA).
In situ hybridization
At first, antisense and sense RNA probes labeled with digoxigenin (DIG) were prepared as described previously (Ref). For hybridization, the tissues were cut into 10-μm thick sections. Using the recommended protocol, we performed in situ hybridization using the DIG-labeling Hybridization Kit (Rosh, USA). Briefly, the tissue sections were hybridized with the DIG-labeled probes using the RiboHybe hybridization solution (Ventana, USA), for 20 h at 60 °C. Then the hybridized samples were washed twice with 1× PBS for 5 min each. After this step, the samples were fixed with 60% formamide followed by another washing step with 0.2× SSC that was carried out 3 times for 5 min each at 37 °C. The slides were then incubated with anti-DIG antibody in blocking solution and NBT/BCIP working solution. Finally, the samples were counter stained using methyl green and the slides were mounted for visualization using the microscope.
Statistical analysis
Real-time PCR results were analyzed for statistical significance using the SAS package (Statistical Analysis System Institute, Version 9.4, Cary, NC, USA). The data are presented as mean ± SD, and the significant difference between groups was determined at P < 0.05.
RESULTS AND DISCUSSION
Paternity analysis and analysis of change in coat color according to the genetic pattern of MC1R
The results obtained by analyzing the change in coat color by crossbreeding based on MC1R genotype analysis are shown in Tables 1, 2, and 3. MC1R genotype results showed that from a total of 325 analyzed KBC, the presence of 164 brindle (50.46%), 65 yellow (20.00%), and 96 black (29.54%) cattle, which belonged to the brindle group corresponded to 52%. According to MC1R genotype analysis by group, out of the 164 cattle identified in the brindle group, 97 of type E+/E+ (62.99%) and 67 of type E+/e (48.20%) were genotyped, and e/e type was not detected, whereas out of 65 cattle identified in the yellow group, 12 of type E+/E+ (7.79%), 21 of type E+/e (15.11%), and 32 of e/e type (49.23%) were genotyped. In case of the black groups (96), 45 of E+/E+ type (29.22%) and 51 of E+/e type (36.69%) were genotyped, and e/e type was not detected. The highest rate of brindle expression was found in 19 (79.17%; E+/E+ type, brindle group) out of 24 brindle off springs that were all obtained from brindle group mother (E+/E+ type) crossed with brindle group father (E+/E+ type). In the case where both the parents had all black coat color, out of the 33 off springs of the mother (E+/e) × father (E+/e type), we obtained 29 (87.88%) black off springs with 10 E+/E+ type (black color group) and 19 E+/e type (black color group).
Expression pattern of genes in MAPK pathway associated with melanogenesis
From the results of melanogenesis-associated gene expression analysis, we identified melanogenesis-associated genes that were differentially expressed in the tissues of the KBC and Korean Hanwoo cattle. MAPK plays a crucial role in the activation of MITF, which further activates the TYR gene. In this study, the brindle cattle highly expressed the FGFR family genes, the main factors of MAPK, compared to that by the Hanwoo cattle. However, the minor genes were highly expressed in Hanwoo. The expression of proteins belonging to Wnt, which inhibits MITF activity, and FZD, which are important active factors, families was higher in Hanwoo than in brindle cattle. Moreover, the expression of the MITF and TYR genes, known as the major active factors of melanogenesis, was significantly higher in brindle cattle than in Hanwoo cattle. These results demonstrated that the cell activity and the main factors of MAPK pathway are highly expressed in brindle cattle. However, the expression of Wnt, which has an opposite effect, was higher in Hanwoo than in brindle cattle (Table 4).
In situ hybridization analysis of MC1R mRNA
The sequence of the exon, in the MC1R gene, that encodes eumelanin was analyzed. The expression of MC1R mRNA in the skin tissue of the Hanwoo (yellow), brindle cattle (brindle and black), and dairy cattle was analyzed and it was revealed that the mRNA expression of MC1R mRNA in the basal layer where melanocytes are distributed was considerably high. Especially, in the brindle cattle group (brindle, black hair), MC1R was highly expressed in the cytoplasm of melanocytes derived from the base of the epidermal layer. On the contrary, dairy cows were found to express MC1R throughout the epidermis. In other words, brindle cattle exhibit very high expression of MC1R in the epidermal basal layer compared to that in other species (Figure 2).
Gene expression patterns of MC1R and TYR in epidermal tissue
By analyzing the protein expression pattern of MC1R and TYR in melanocytes, keratinocytes, and epidermis sections of brindle and black hairs of Hanwoo and brindle cattle, we observed that the MC1R and TYR protein expression levels were very low in Hanwoo cattle (yellow hair) but were strongly expressed in brindle and black cattle tissues (Figure 3). In particular, the expression of these proteins in melanocytes and epidermis was high in brindle and was rather low in black cattle. However, TYR protein expression was higher than that of MC1R in both, melanocytes and keratinocytes of Hanwoo cattle. On the contrary, in brindle cattle, the expression level of TYR protein was low in melanocytes but was high keratinocytes. In general, in the black cattle, the expression of TYR protein in keratinocytes was high and the expression in melanocytes was lower than that in the brindle cattle. These results were similar to those of the expression pattern of MC1R and TYR at the mRNA level. The expression of MC1R and TYR in melanocytes was higher than that in keratinocytes of brindle cattle, but in the case of black cattle, it was higher in keratinocytes than in melanocytes. Therefore, the results demonstrate that the expression of MC1R was low and the expression of TYR was high in yellow and brindle tissues except for black tissue. In other words, the reaction of the pigment deposition increases in the hair because the expression of TYR was higher than the expression of MC1R in the keratinocytes.
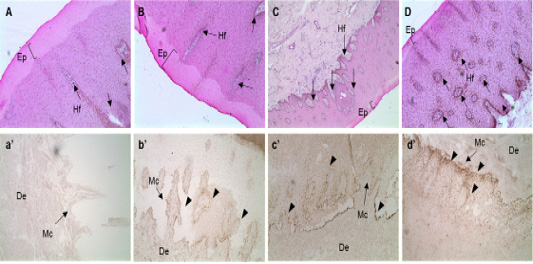
Figure 2: In situ hybridization of MC1R mRNA in the follicular zone of cattle. Black bar = 100 μm. Black arrows indicate MAP1LC3A RNA probe detection. MC1R probe was made using NCBI genetic information EU169234, sequence is “ctgtgtctga cttgctggtg agcgtcagca acgtgctgga gacggcagtc atgccgctgc tggaggccgg tgtcctggcc acccaggcgg ccgtggtgca gcagctggac aatgtcatcg”. A, B, C and D: H&E staining of skin tissues of cattle. a’, b’, c’ and d’: in situ hybridization of MC1R probe detection dots. *, ** Different letters within the same column represent a significant difference (p<0.05). A-a’) Hanwoo (HC), B-b’) Korean brindle cattle, C-c’) Korean dark cattle, D-d’) Holstein cattle. Hf: Hair follicle, Ep: epidermis, Mc: Melanocyte zone, De: dermis. A–D: 100X magnification; a’-d’: 200X magnification.
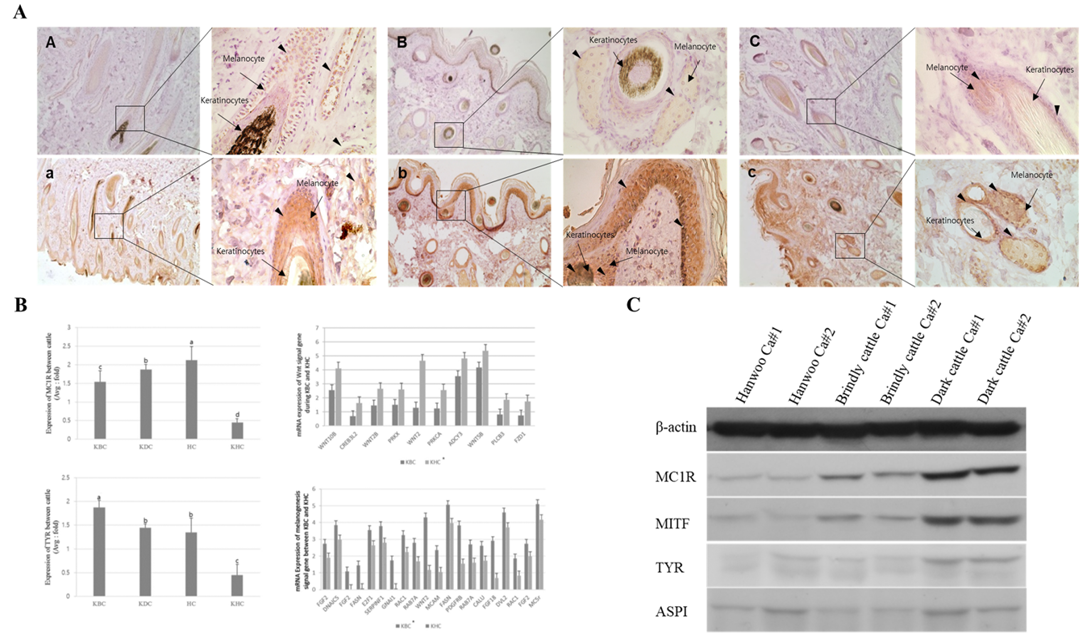
Figure 3: Expression analysis of melanogenesis-associated genes between the Korean native cattle. A: Immunodetection of MC1R and TYR protein A-D) Detection of MC1R protein, a-d) Detection of TYR protein, A-a) Hanwoo, B-b) Korean brindle cattle, C-c) Korean dark cattle. B: Real-time qPCR analysis of melanogenesis-associated genes, where KBC) Korean brindle cattle, KDC) Korean dark cattle, HC) Holstein cattle, KHC) Korean Hanwoo cattle. The bars represent the average fold changes in 3 independent experiments (±SD). C: Western blot analysis.
Table 2: MC1R genotype and coat color in sire (E+/E+ , E+/e) and dam (E+/E+) of Korean brindle cattle.
| Dam | Sire | Offspring | % | Dam | Sire | Offspring | % | ||||||||
| MC1R | Coat | MC1R | Coat | MC1R | Coat | No. | MC1R | Coat | MC1R | Coat | MC1R | Coat | No. | ||
| E+/E+ | A group | E+/E+ | A group | E+/E+ | A | 19 | 79.17 | E+/E+ | A group | E+/e | A group | E+/E+ | A | 24 | 42.11 |
| B | 2 | 8.33 | B | 7 | 12.28 | ||||||||||
| C | 3 | 12.50 | C | 6 | 10.53 | ||||||||||
| E+/e | A | E+/e | A | 12 | 21.05 | ||||||||||
| B | B | 6 | 10.53 | ||||||||||||
| C | C | 2 | 3.51 | ||||||||||||
| e/e | A | e/e | A | ||||||||||||
| B | B | ||||||||||||||
| C | C | ||||||||||||||
| Total | 24 | 100 | Total | 57 | 100 | ||||||||||
Table 3: MC1R genotype and coat color in sire (E+/e) and dam (E+/E+ , E+/e , e/e) of Korean brindle cattle.
| Dam | Sire | Offspring | % | Dam | Sire | Offspring | % | ||||||||
| MC1R | Coat | MC1R | Coat | MC1R | Coat | No. | MC1R | Coat | MC1R | Coat | MC1R | Coat | No. | ||
| E+/E+ | C group | E+/e | C group | E+/E+ | A | 0 | 0.00 | E+/e | C group | E+/e | A group | E+/E+ | A | 2 | 20.00 |
| B | 0 | 0.00 | B | 1 | 10.00 | ||||||||||
| C | 7 | 50.00 | C | 1 | 10.00 | ||||||||||
| E+/e | A | 0 | 0.00 | E+/e | A | 4 | 40.00 | ||||||||
| B | 0 | 0.00 | B | 0 | 0.00 | ||||||||||
| C | 7 | 50.00 | C | 1 | 10.00 | ||||||||||
| e/e | A | 0 | 0.00 | e/e | A | 0 | 0.00 | ||||||||
| B | 0 | 0.00 | B | 1 | 10.00 | ||||||||||
| C | 0 | 0.00 | C | 0 | 0.00 | ||||||||||
| Total | 14 | 100 | Total | 10 | 100 | ||||||||||
| Dam | Sire | Offspring | % | Dam | Sire | Offspring | % | ||||||||
| MC1R | Coat | MC1R | Coat | MC1R | Coat | No. | MC1R | Coat | MC1R | Coat | MC1R | Coat | No. | ||
| E+/e | C group | E+/e | C group | E+/E+ | A | 0 | 0.00 | e/e | B group | E+/e | A group | E+/E+ | A | 0 | 0.00 |
| B | 0 | 0.00 | B | 0 | 0.00 | ||||||||||
| C | 10 | 30.30 | C | 0 | 0.00 | ||||||||||
| E+/e | A | 0 | 0.00 | E+/e | A | 11 | 52.38 | ||||||||
| B | 0 | 0.00 | B | 2 | 9.52 | ||||||||||
| C | 19 | 57.58 | C | 3 | 14.29 | ||||||||||
| e/e | A | 0 | 0.00 | e/e | A | 0 | 0.00 | ||||||||
| B | 4 | 12.12 | B | 5 | 23.81 | ||||||||||
| C | 0 | 0.00 | C | 0 | 0.00 | ||||||||||
| Total | 33 | 100 | Total | 21 | 100 | ||||||||||
According to a report on the current status of livestock genetic resources published by the Ministry of Agriculture and Forestry, the determination of coat color is an important method for classifying the domestic animals. Thus, the phenotype of domestic livestock can be reliably used to assign value to the natural genetic resources and to identify the species. Unlike the Hanwoo cattle, which has yellow coat color and the Jeju Black cattle, which has typical black coat color, the brindle cattle exhibits three distinct coat color phenotypes, including yellow, brindle, and black, which varies depending on the local tendencies and breeding environment. Lee et al. (2011) reported that
Table 4: Melanogenesis-associated genes that are up-regulated and down-regulated in the KBC and KHC.
| Probe ID | Gene symbol | Description | Primary accession | KBC | HW |
| A_73_112907 | ADCY2 | PREDICTED: Bos taurus adenylate cyclase 2 | XM_587884 | 1.8571751 | -2.4811394 |
| A_73_P082846 | ADCY3 | PREDICTED: Bos taurus adenylate cyclase 3 | ENSBTAT00000032950 | -4.812259 | -3.5481966 |
| A_73_104723 | CREB3L2 | Bos taurus cAMP responsive element binding protein 3-like 2 | NM_001102533 | 0.63402313 | -1.6985123 |
| A_73_100378 | FGF13 | Bos taurus fibroblast growth factor 13 | NM_001098892 | 0.18493223 | -3.783594 |
| A_73_117785 | FGF16 | Bos taurus fibroblast growth factor 16 | NM_001192777 | -2.6347258 | -4.695286 |
| A_73_102234 | FGF18 | Bos taurus fibroblast growth factor 18 | NM_001076007 | -2.6800463 | -1.1037806 |
| A_73_100047 | FGF2 | Bos taurus fibroblast growth factor 2 | NM_174056 | -3.9870496 | -2.7472577 |
| A_73_P050691 | FGFR1 | Bos taurus fibroblast growth factor receptor 1 | NM_001110207 | 2.6757064 | 3.846626 |
| A_73_P258736 | FZD1 | Bos taurus frizzled homolog 1 | NM_001101048 | -1.7578547 | -0.754021 |
| A_73_P258696 | GNAO1 | Bos taurus guanine nucleotide binding protein (G protein) | NM_174325 | -3.730153 | -4.9742236 |
| A_73_P032176 | MITF | Bos taurus microphthalmia-associated transcription factor (MITF) | NM_001001150 | 0.562865995 | -1.7221494 |
| A_73_113415 | PIK3R2 | Bos taurus phosphoinositide-3-kinase, regulatory subunit 2 | NM_174576 | 1.502133 | 2.6541512 |
| A_73_P036956 | MC1R | Bos taurus melanocortin 1 receptor ( (MC1R) | NM_174108 | 1.321908 | 0.402622 |
| A_73_P037571 | TYR | Bos taurus tyrosinase (TYR) | NM_181001 | 5.273953 | 2.398885 |
| A_73_P040651 | DCT | Bos taurus dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) | NM_001012666 | 9.706837 | 3.279001 |
| A_73_100318 | MAPK14 | Bos taurus mitogen-activated protein kinase 14 (MAPK14) | NM_001102174 | 0.734054 | -0.44604 |
| A_73_105507 | MAPK13 | Bos taurus mitogen-activated protein kinase 13 (MAPK13) | NM_001014947 | 0.732774 | -0.44856 |
| A_73_P044751 | RAF1 | Bos taurus v-raf-1 murine leukemia viral oncogene homolog 1 (RAF1)] | NM_001102505 | 0.924042 | -0.11397 |
| A_73_P257606 | MAP2K1 | Bos taurus mitogen-activated protein kinase kinase 1 (MAP2K1) | NM_001130752 | 03988285 | -0.017 |
| A_73_P411006 | TYRP1 | Bos taurus tyrosinase-related protein 1 (TYRP1) | NM_174480 | 2.182334 | 1.125872 |
| A_73_P109681 | PLCB3 | Bos taurus phospholipase C, beta 3 | NM_001192423 | -1.8584734 | -0.824501 |
| A_73_P342431 | POMC | Bos taurus proopiomelanocortin | NM_174151 | 0.504154 | -0.5090313 |
| A_73_P066661 | PRKCA | Protein kinase C alpha type | ENSBTAT00000001407 | -2.5464063 | -1.2484481 |
| A_73_119927 | PRKX | PREDICTED: Bos taurus protein kinase, X-linked | ENSBTAT00000019102 | -2.6099973 | -1.5094379 |
| A_73_105798 | TYRP1 | Bos taurus tyrosinase-related protein 1 | NM_174480 | -1.3673902 | -3.7579913 |
| A_73_102442 | WNT10B | PREDICTED: Bos taurus wingless-type MMTV integration site family, member 10B | XM_586498 | -4.114833 | -2.5609531 |
| A_73_P051501 | WNT2 | Bos taurus wingless-type MMTV integration site family member 2 | NM_001013001 | -4.659621 | -1.3054186 |
| A_73_108402 | WNT2B | Bos taurus wingless-type MMTV integration site family, member 2B | NM_001099363 | -2.6472654 | -1.4541558 |
| A_73_P091651 | WNT5B | PREDICTED: Bos taurus wingless-type MMTV integration site family, member 5B | ENSBTAT00000001766 | -5.3642144 | -4.1710067 |
| A_73_114435 | PDGFC | platelet derived growth factor C | ENSBTAT00000061070 | -4.129871 | -2.8084745 |
out of the 276 brindle cattle that were managed by the Agricultural Technology Center in Ulleungdo, 63%, 24%, and 13% were identified to possess brindle, yellow, and black coat color, respectively. They also revealed that out of the 40 cattle that were managed at the Livestock Technology Research Center in Gangwon-do, 66%, 13%, and 21% were identified to possess brindle, black, and yellow colored coat, respectively. Although the phenotype distribution of the brindle cattle differs between regions, it is shown that, upon analyzing the coat color, the average expression of the brindle is 65%, the black is 13%, and the yellow is 22%. From the brindle cattle experiment in this study, approximately 56% exhibited the brindle color, and thereafter, more than 50% of the brindle color phenotype was observed in brindle cattle, which is similar to the above indicated studies, with 20% of black and 24% of yellow colored cattle. The expression of diverse coat color, which is dependent on the genetic variation of MC1R, known as the top-level modulator of eumelanin or pheomelanin synthesis, that are synthesized in melanocytes between the dermal and epidermal layers, plays an important role in not only the coat color decision of this generation but also the coat color decision of the later generations (Klungland et al., 1995). The three alleles of MC1R (ED, E+, e), which regulate the genetic expression of the pigmentation-related genes in Hanwoo, E+ operates as a normal receptor in the MSH and melanocyte cell membranes and is known to cause pigmentation such as black or red by other mechanisms associated with melanogenesis. For e genotypes, it is also known that the chromosomes possess frameshift mutation that leads to red pigmentation with homomorphism (Klungland et al., 1995; Adalsteinsson et al., 1995). However, in the case of Hanwoo that expresses yellow color, the genetic variation of e/e, which is molded with e allele, was the main reason. Previously, it was shown that expressions of E+/e and E+/E+ appears only in the case of brindle (Kim et al., 2013). In the study conducted by Lee et al. (2000), the MC1R locus on the black-colored phenotype of Jeju Black cattle reported in the formation of the black-colored hair by the gene action of E+/- and Agouti allele was also found to played a role. In the study conducted by Lee et al. (2002), the variability of MC1R for coat color phenotypes of brindle cattle was identified to be due the genotypes of E+/E+ and E+/e, and the expression of e/e was not observed, which suggested that the pigmentation of the coat color by E+ could be responsible for the brindle-colored phenotype. The results of this study show that E+/E+ or E+/e are the genotypes of the individuals that show the brindle color, unlike previous results (Lee et al., 2002), and the yellow-colored individuals with the e/e genotype were also obtained in the offspring of parents who had the genotype of E+/e. This result suggests that the genotype variations in MC1R might be different for the coat color in the brindle cattle. However, in the normal mechanism of α-MSH, brindle cattle of yellow phenotype showed the same genotype as that previously reported (Robbins et al., 1993; Jackson et al., 1993; Klungland et al., 1995) leading to the red color phenotype induced by MC1R, which is expressed by the e/e homogenous junction, and it does not involve any signal transduction of melanogenesis mechanism in the cytoplasm of the pigmented cells. The antagonistic aspect of the phenotype of the MC1R gene in cattle was also reported to affect the combination of α-MSH with heterozygotes or homozygotes (Chung et al., 2000; Lee et al., 2002). These variations can also play very important role in the gene expression of TYR, TYRP1, and DCT for synthesizing melanin. However, these variations are introduced by genetic processes or the DNA methylation of epigenome in MC1R, and these mechanisms are demonstrated to play an important role in the synthesis of melanin and the final melanin metabolite (Agar et al., 2005). Thus, in this study, we analyzed whether the change in the coat color in brindle cattle and the variations in related genes have a characteristic association with the melanogenesis mechanism, which might further depend on the type of pigmentation induced within the melanocytes of brindle cattle. The melanocyte metabolic reactions affect the cell cycle activity of cells in brindle cattle, which is based on the differences in the coat color and it has been confirmed in a report by Kim et al. (2014). In addition, the MC1R expression levels, according to the coat color phenotype were high in black and brindle color groups, excluding the yellow colored group, and the expression levels of TYR and DCT, which induce melanin synthesis, followed the same pattern. However, comparing the gene expression results between black and brindle color groups, that in the black color group was significantly higher. In particular, in the highly expressed brindle color group, the expression of MAPK and Wnt genes that are related to the mechanisms of MC1R and melanogenesis was significant high in the brindle-colored pigment cells. Moreover, the expression levels of TYR and DCT, which directly affect the activity of tyrosine, were also increased. In other words, the expression of a majority of the Wnt signaling pathway genes can be thought of having a small role in regulating the pigment synthesis and the pigmentation rate in colored cells. However, it was observed that melanin synthesis was inhibited due to the negative effect on the expression of genes related to melanin pigment activity. Moreover, the result suggests that MAPK-related genes enhance the expression of the factors that affect cell cycle activity or regulate the expression of the factors that inhibit cell cycle leading to an increase in cell cycle activity. The expression of MC1R and other major genes was significantly increased in the brindle cells, which could be attributed to the reduced action of the mechanisms associated with MMPs and inhibition of cell cycle activity. Same results, as those obtained in this study, were reported previously showing that cell division and mechanism of melanin synthesis was activated (Vage et al., 1999; Klungland et al., 1995). The analysis of protein expression pattern suggests that the expression of MITF that activates TYR and DCT in the brindle color group, which is also associated to an increase in the expression of MC1R gene, was significantly increased compared to other coat colors. These results are similar to the one presented in the study conducted by Kim et al. (2013) which showed that higher expression of MAPK-related genes in the case of brindle-colored cells leads to an increase in brindle color and black color. The high expression of Wnt-associated genes was also thought to be able to inhibit cell cycle activity and the expression of melsanogenesis-related genes in yellow cattle.
CONCLUSIONS AND RECOMMENDATIONS
Thus, the results of this study have revealed that the coat color expression of KBC is different from that of the ordinary yellow colored cattle. Therefore, we believe that the KBC should be classified as a different breed among Korean native cattle, and we further suggest that the results of this study can be used as a measure to identify and differentiate the KBC from other native cattle.
Novelty Statement
This study was able to demonstrate the diversity of native livestock by evaluating mutations and expression differences in melanogenesis-related genes, particularly MC1R or TYR.
AUTHOR’S CONTRIBUTION
Kyoung Sub Jung; Investigation, Experiment, Data analysis, Formal analysis. Kyoung Sub Jung, Yun Dong Choi; Experiment, Statistical analysis, Methodology, Writing of Original draft. Sang Hwan Kim; Editing, Review, Supervision.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES






