South Asian Journal of Life Sciences
Research Article
Proteomic Profiling of Freeze- and Spray-Dried Water Extracts of Snakehead Fish (Channa striatus): In Search of Biomolecules for Wound Healing Properties
Soon Hong Kwan1, Saringat Baie2, Nornisah Mohammed2, Mohd Nazri Ismail1*
1Doping Control Centre, Universiti Sains Malaysia, 11800, USM, Penang, Malaysia; 2School of Pharmaceutical Sciences, Universiti Sains Malaysia, 11800, USM, Penang, Malaysia.
Abstract | Channa striatus is a carnivorous freshwater fish that is commonly consumed among Malaysians. The fish is known to contain compound(s) that can accelerate the wound healing process in humans, but the attributes of these compound(s) are yet to be clarified. In the current study, we have performed a thorough proteomic profiling of spray -dried and freeze-dried C. striatus water extracts using high-sensitivity liquid chromatography tandem mass spectrometry. Other than the analysis of whole sample, both samples were also fractionated in order to maximise protein detection. About 137 and 194 proteins were identified in spray dried and freeze dried samples, respectively. Actin, myosin, tropomyosin, calcium ion-related protein and collagen are among the proteins that have been identified, and which are suspected to be involved in the wound healing process. A high number of uncharacterised proteins were also detected, which suggested that there are still many fish proteins with unknown functions in C. striatus. In the future, the identified proteins can be isolated and further studies are required for a better understanding on the wound healing property of C. striatus.
Keywords | Channa striatus, Fish, Mass spectrometry, Proteomics, Wound healing
Editor | Muhammad Nauman Zahid, Quality Operations Laboratory, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | May 20, 2015; Revised | September 27, 2015; Accepted | September 29, 2015; Published | December 17, 2015
*Correspondence | Mohd Nazri Ismail, Doping Control Centre, Universiti Sains Maalaysia, 11800, USM, Penang, Malaysia; Email: [email protected]
Citation | Kwan SH, Baie S, Mohammed N, Ismail MN (2015). Proteomic profiling of Freeze- and spray-dried water extracts of Snakehead fish (Channa striatus): In search of biomolecules for wound healing properties. S. Asian J. Life Sci. 3(1): 22-41.
DOI | http://dx.doi.org/10.14737/journal.sajls/2015/3.1.22.41
ISSN | 2311–0589
Copyright © 2015 Kwan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Channa striatus, is a carnivorous freshwater fish that is commonly found in Malaysia and is commonly known by the locals as “Haruan” in the Malay language. It is the most widely introduced snakehead fish under the family of Channidae (Courtenay and Williams, 2004). Other countries where we can encounter such species include the subtropical countries such as China, Taiwan, Thailand, Philippines, Indonesia and India (Mohsin and Ambak, 1983). Although currently such snakehead fish is cultured commercially in Thailand, Philippines and India (Gam et al., 2006), it was not given priority in the local farming industry as the locals believe it to be a pest (Jais, 2007).
C. striatus has long been a recommended part for the patients’ diet that have just gone through surgery or delivery as the locals believe that it helps in the healing process (Jais et al., 1994). C. striatus flesh is thought to contain medicinal properties and its consumption helps in the recovery from serious illness (Wee, 1982). Such theory has been supported by Jais et al. (1994), who stated that C. striatus contains essential amino acids for wound healing. Jais et al. (1994) also showed a high amount of arachidonic acid in C. striatus. Arachidonic acid is the precursor of thromboxane and prostacyclin that is responsible for blood clotting (Nelson et al., 1997; Silver et al., 1973). In previous studies, a C. striatus extract has been applied on wounded mice to determine the tensile strength of the healed wound (Baie and Sheikh, 2000a, 2000b). They reported that the high content of arachidonic acid, glycine and polyunsaturated fatty acid may have contributed to the wound healing process.
The pharmaceutical products of C. striatus that are currently available in the market are usually in capsule form. C. striatus capsules are prepared by either freeze-drying or spray- drying the whole fish including the water extract. The water extract of the fish actually resembles the traditional preparation of the fish for consumption which is the soup. Both the freeze-drying (Mellor, 1978) and spray-drying (Masters, 1979) methods have their own strengths and weaknesses, which can give impacts on the protein contents retained in the end products. Both of the methods have always been compared using different samples (Anwar and Kunz, 2011; Chen et al., 2012; Maa et al., 1999) in order to identify the better option for each sample respectively. However, until currently both the freeze-dried and spray-dried samples of C. striatus have never been compared. In this study, we have compared the proteomes detected for both samples.
With the advancement of technology, proteins available in the fish can be now be better profiled (Doherty et al., 2012; Martyniuk and Denslow, 2012). Fish are rich in proteins that are essential and beneficial to human health. A better understanding of the interaction between the proteins in the C. striatus and the wound can be established through the profiling of those proteins. In addition, the proteins responsible for the wound healing effect can be isolated for medicinal and clinical purposes once identified. Despite having a preliminary protein profiling being conducted previously on the flesh of C. striatus (Gam et al., 2006), there were still limitations on the profiling as the equipment used in the past were not as sensitive.
In this study, we have compared the protein concentration and performed proteome profiling for both freeze-dried and spray-dried water extracts from C. striatus. Other than analysing the whole sample, we have also analysed fractionated samples to complement the result. The aim was to profile as many proteins as possible and postulate those that might be involved in the wound healing property of C. striatus. Such findings would give us an insight on how the consumption of C. striatus could contribute to the wound healing mechanism.
Materials and methods
Protein Extraction from C. striatus Samples
The C. striatus freeze- and spray-dried samples were provided by the School of Pharmaceutical Sciences, Universiti Sains Malaysia. The C. striatus originated from Kedah, Peninsular Malaysia. Protein extraction on both samples was performed in replicates according to the method reported by Gam et al. (2006) with minor adjustments. 15mg of freeze-dried and spray-dried C. striatus sample were added into 1 mL of 40 mMTris-HCl (pH 8.8) extraction buffer (Bio-Rad Laboratories, CA, USA) respectively and waited for 20 minutes with occasional vortex. Sample mixtures were then centrifuged at 12,000 x g for 30 minutes. Supernatants were recovered and kept in -35°C for subsequent analysis.
Total Protein Quantification using Bradford Assay
Total protein quantification for the sample of C. striatus was carried out according to Bradford (1976). 5µL of supernatant collected during the protein extraction was mixed with 250µL of Bradford reagent in a 96 well plate. The solution was then incubated for 15 minutes in room temperature. The absorbance was then measured at 595 nm. A standard curve was constructed using the bovine serum albumin (BSA). The standard curve plotted ranged from 0.0-1.4 mg/mL. The total protein concentration in each sample was determined by comparing the absorbance value obtained for the sample against the standard curve.
Protein Fractionation using Gelfree 8100 Fractionation System
The procedure was carried out on Gelfree 8100 fractionation system (Expedeon, CA, USA) according to the article by Witkowski and Harkins (2009). 200 µg of protein samples were loaded to both 8% Tris-acetate cartridge and 10% Tris-acetate cartridge. Twelve fractions were collected during the procedure, respectively. In order to confirm that the separation was successful, 10 µl of each fraction was analysed using SDS-PAGE (12.5% gel) (Bio-Rad Laboratories, CA, USA) and stained with Coomassie blue. The remaining fractions were then concentrated using a concentrator to remove the sample buffer.
Protein Digestion using Trypsin
The protein samples were re-suspended in 100µL of 6 M urea, 100mMTris buffer at 10mg/mL. The digestion method was carried out according to Kinter and Sherman (2005). Briefly, 200 mM DTT was added to each sample and kept in room temperature for 1 hour. Later, 200 mM of iodoacetamide was added and incubated in room temperature for 1 hour. 20 µL of 200 mM DTT was added next to consume any unreacted iodoacetamide. Concentration of urea in the sample was then reduced by adding 775µL of water. 20µg of trypsin (Promega, WI, USA) in solution is added to each sample and incubated overnight at 37 °C for digestion purpose. The digestion was stopped the next day by adjusting the pH of the buffer to pH <6.
LC-MS/MS Analysis
Prior to LC-MS/MS analysis, each of the peptide sample were mixed with 100ul of 0.1% formic acid in deionized water and filtered using the 0.45um regenerated cellulose (RC) membrane syringe filter (Sartorius AG, Goettingen, Germany). Analysis was performed using LTQ-Orbitrap Velos Pro mass spectrometer coupled with Easy-nLC II nano liquid chromatography system. Easy column C18 (10cm, 0.75mm i.d., 3μm; Thermo Scientific, San Jose, CA, USA) was used as the analytical column, whereas Easy column C18 (2cm, 0.1mm i.d., 5μm; Thermo Scientific, San Jose, CA, USA) was used as the pre-column. The pre-column was equilibrated at a flow rate of 3μL/min for 15μL and analytical column was equilibrated at a flow rate of 0.3μL/min for 4μL. 5μL of samples were injected and chromatographically separated at a flow rate of 0.3μL/min. Running buffers used were: (A) 0.1% formic acid in deionized water, and (B) 0.1% formic acid in acetonitrile. Samples were eluted using the gradient 5% to 100% of buffer B in 80 minutes. The eluent was sprayed into the mass spectrometer at 2.1 kV (source voltage) and capillary temperature of 220˚C was used. Peptides were detected by full scan mass analysis from m/z 300-2,000 at resolving power of 60,000 (at m/z 400, FWHM; 1-s acquisition), with data-dependent MS/MS analyses (ITMS) triggered by the 8 most abundant ions from the parent mass list of predicted peptides, with rejection of singly or unassigned charge state. Collision induced dissociation (CID) was applied as the fragmentation technique with a collision energy of 35. Each sample was analysed twice.
Protein and Peptide Identification by De Novo Sequencing
PEAKS Studio Version 7 (Bioinformatics Solution, Waterloo, Canada) was used to perform de novo sequencing and database matching. Uniprot fish database from October 2014 was used for the database matching. Carbamidomethylation and methionine oxidation were set as fixed modifications and maximum missed cleavage was set at 2. Parent mass and precursor mass tolerance were set at 0.1 Da. False detection rate (FDR) <0.1% and significant score (−10lgP) for protein >30 were used for protein acceptance. Maximum variable post-translational modification was set at 4.
Results and Discussion
Total Protein Concentration
The total protein concentration varies for each C. striatus sample. The average result from four replicates is shown in Table 1. As presented, the total protein concentration in the spray-dried sample is slightly lower than the freeze-dried sample. This might be due to the high temperature introduced during the spray-drying process, which has caused denaturation on certain proteins to be available in the sample. Joshi et al. (2011) has mentioned that different drying methods have different capabilities of denaturing proteins by modifying the protein structures. Figure 1, which is a SDS-PAGE image, showed the application of Gelfree fraction system able to separate proteins in the C. striatus samples. The fractionated proteins allow the mass spectrometry to detect low abundance proteins available in the samples.
Table 1: Average total protein concentration in C. striatus free-dried and spray-dried samples
|
Sample |
Average total protein concentration (mg/ml) |
|
Freeze-dried (FD) water extracts |
1.242 |
|
Spray-dried (SD) water extracts |
1.160 |
12 fractions were collected from freeze-dried and spray-dried samples, respectively.
Many proteins are heat-sensitive and thus spray-drying has been less favourable in the pharmaceutical industry. During the spray-drying process, the denaturation temperature of protein is greatly affected by the water content in the sample (Maltesen and Van De Weert, 2008). Proteins are most sensitive to thermal denaturation after atomisation. Such phenomenon gives an indication that proteins face a higher risk of denaturation in spray-drying method. It explains the reason for the lower protein concentration obtained in the spray-drying sample. Therefore, the pharmaceutical industry and research work prefer the freeze-drying method (Maltesen and Van De Weert, 2008), whereas the food industry tends to select the spray-drying method.
Comparison of Proteome Profiles Obtained from Freeze- and Spray-Dried C. striatus Water Extracts
Protein identification by nano-LCMS/MS showed that 137 proteins and 194 proteins were identified in spray-dried and freeze-dried samples respectively. The amount of proteins shown consists of the accumulation of whole sample as well as the fractions obtained from Gelfree fractionation. The number of proteins identified is higher compared to the previous proteomic analysis by Gam et al. (2006), which only obtained 85 proteins, which suggested that the utilization of advanced Easy-nLC II nano liquid chromatography system enables more minor proteins to be detected. Such result was in line with the total protein concentration obtained, where the freeze dried sample is higher. A portion of proteins in the C. striatus sample that are heat-sensitive might have been denatured during the spray drying process. 97 of the proteins identified in the freeze-dried sample were similar to the proteins identified in the earlier spray-dried sample as shown in Figure 2. The similar proteins were mainly consisted of structural proteins and uncharacterised proteins.
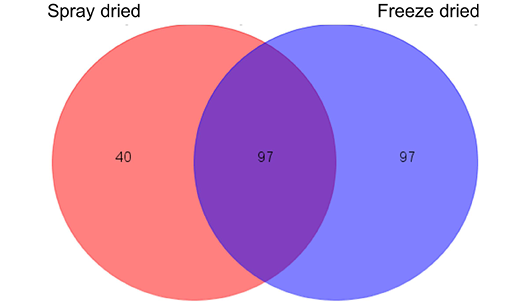
Figure 2: Venn diagram of protein numbers detected in both spray-dried (left) and freeze-dried (right) C. striatus water extracts
A number of 97 identical proteins were detected on both samples.
Role of Proteins in Wound Healing
In general, three different but overlapping phases can be identified in the wound healing process which is: (1) haemostasis and inflammation, (2) proliferation or granulation, and (3) maturation or remodelling (Flanagan, 2000; Singer and Clark, 1999; Witte and Barbul, 1997). Different proteins are involved and very often they are integrated into a complex mechanism. Several proteins that were identified in the C. Striatus could give us an insight on how it enhances the wound healing mechanism. The list of proteins detected for both samples were listed in Table 2 and Table 3. Due to the limited entries in the protein database for C. striatus, a large amount of proteins detected from the database were from different species other than C. striatus. This also showed the possibility that the proteins/peptides available in C. striatus share the same sequence with other fish species as well.
Actin, Myosin and Tropomyosin
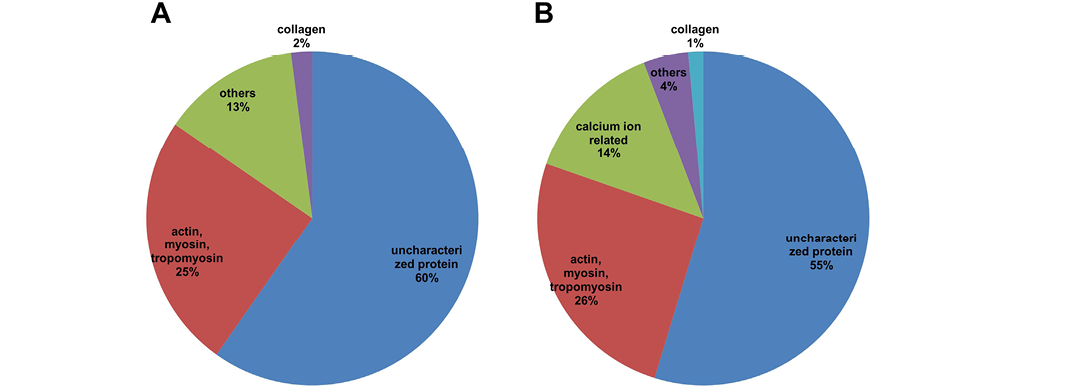
Actin, myosin and tropomyosin constitute 25% and 26% of the total protein detected in freeze- and spray-dried C. striatus water extracts, respectively (Figure 3). According to Dominguez and Holmes (2011), actins are essential for cellular functions such as motility, the maintenance of cell shape and polarity, cell division and cytokinesis, vesicle and organelle movement, cell signalling as well as the establishment and maintenance of cell junctions, and regulation of transcription. It often takes place during the re-epitheliasation, where it is part of the proliferation phase. A report by Martin and Lewis (1992) mentioned that when embryonic wound occurred, the actin cable plays the role to close up the embryonic wound. The rapidly assembled actin purse string is responsible for the closure of the wound in the embryonic epidermis (Richardson et al., 2013). Therefore, actin plays an important role in embryogenesis as well as in wound healing, where cell motility is crucial for the sealing of wound margins. Other structural proteins also play a role in the remodelling of the tissue structure at the wound area. Actin works together with myosin in the wound healing process. The biomechanical process driving cell motility and relation between actin and myosin has been clearly demonstrated by Giannone et al. (2007). Conrad et al. (1993) published a report supporting that actin, myosin I and myosin II were involved in the wound healing process of fibroblasts. Myosin II can be regulated in order to accelerate the healing of large wounds and thus improving a wound in healing (Matsubayashi et al., 2011). Tropomyosin is a multi-isoform family of actin-associating proteins that controls isoform-specific regulation of diverse actin filaments (Bach et al., 2009; Gunning et al., 2005). A recent report by Lees et al. (2013) suggested that
A: freeze-dried sample; B: spray-dried sample; The proteins were categorized into 5 groups: uncharacterised proteins; actin, myosin, tropomyosin; calcium ion-related proteins; collagen; others.
the tropomyosin may be important regulators of actin functioning during the wound healing process. In short, actin, myosin and tropomysosin are directly involved in the regulation and maintenance of wound recovery. Therefore, the high amount of myosin, actin and tropomyosin may give us the insight of the ability of accelerated wound healing properties promoted by the C. striatus application.
Calcium Ion-Related Protein
Another interesting finding obtained was related to the calcium ion-related protein detected in the samples. Ca2+ is known to play a role in directing cell polarity and guiding highly polarized processes like tip growth (Messerli and Robinson, 2007), neurite extension (Zheng and Poo, 2007), and cell migration (Maroto and Hamill, 2007). A research finding published by Graham et al. (2013) revealed that calcium signalling plays a role in inducing the cellular organization and also the migration of skin cells. The introduction of calcium ion would trigger the local fusion of internal membranes, creating a patch vesicle that staunches the wound (McNeil and Kirchhausen, 2005). However, Kono et al. (2012) has highlighted that the calcium ion entry has to be well-regulated because it is lethal in large amounts, but is required at moderate levels to induce the wound healing process. Such observation emphasized that calcium ion-related protein plays a significant role in regulating and initiating the process of wound healing once the wound is inflicted. However, the calcium transporting enzyme is only detected in spray-dried sample (Table 3) but not in the freeze-dried sample. The reason of such phenomenon is not clear and requires more extensive study.
Table 2: List of detected proteins in freeze-dried C. striatus samples from a combination of whole sample and fractions analysis
|
Accession |
Max -10lgP |
(%) Max coverage |
Max # peptides |
Max # unique |
Description |
|
tr|H2LNX8|H2LNX8_ORYLA |
321.96 |
44 |
50 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=MYH13 (4 of 11) PE=4 SV=1 |
|
tr|Q05K09|Q05K09_ORYLA |
320.56 |
23 |
50 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-2 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q05K05|Q05K05_ORYLA |
318.51 |
23 |
52 |
2 |
Fast skeletal myosin heavy chain isoform mMYH-11 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q05K11|Q05K11_ORYLA |
312.54 |
22 |
45 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-5 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q05K06|Q05K06_ORYLA |
312.12 |
22 |
45 |
0 |
Fast skeletal myosin heavy chain mMYH-9 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q05K07|Q05K07_ORYLA |
310.55 |
23 |
46 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-7 OS=Oryzias latipes PE=4 SV=1 |
|
tr|G0YU50|G0YU50_SINCH |
308.34 |
18 |
40 |
2 |
Fast skeletal muscle myosin heavy chain isoform 3 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|G0YU48|G0YU48_SINCH |
308.34 |
18 |
40 |
2 |
Fast skeletal muscle myosin heavy chain isoform 1 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|G0YU49|G0YU49_SINCH |
308.34 |
18 |
40 |
2 |
Fast skeletal muscle myosin heavy chain isoform 2 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|Q6SNT2|Q6SNT2_SINCH |
308.34 |
18 |
40 |
2 |
Fast skeletal muscle myosin heavy chain OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|H2LPK6|H2LPK6_ORYLA |
307.58 |
20 |
45 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=MYH13 (7 of 11) PE=4 SV=1 |
|
tr|Q05K10|Q05K10_ORYLA |
307.16 |
20 |
45 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-1 OS=Oryzias latipes PE=4 SV=1 |
|
tr|H2N1T3|H2N1T3_ORYLA |
305.63 |
22 |
40 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=MYH13 (11 of 11) PE=4 SV=1 |
|
tr|H2LPD7|H2LPD7_ORYLA |
305.52 |
20 |
39 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=MYH13 (6 of 11) PE=4 SV=1 |
|
tr|C4TIP2|C4TIP2_ORYLA |
304.90 |
17 |
36 |
0 |
Myosin heavy chain larval type 1 OS=Oryzias latipes GN=mMYHL1 PE=2 SV=1 |
|
tr|Q05K12|Q05K12_ORYLA |
304.48 |
21 |
42 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-6 OS=Oryzias latipes PE=4 SV=1 |
|
tr|C4TIP1|C4TIP1_ORYLA |
302.57 |
15 |
37 |
0 |
Myosin heavy chain embryonic type 1 OS=Oryzias latipes GN=mMYHemb1 PE=2 SV=1 |
|
tr|Q05K08|Q05K08_ORYLA |
302.01 |
19 |
40 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-3 OS=Oryzias latipes PE=4 SV=1 |
|
tr|C4TIP3|C4TIP3_ORYLA |
301.82 |
16 |
35 |
0 |
Myosin heavy chain larval type 2 OS=Oryzias latipes GN=mMYHL2 PE=2 SV=1 |
|
tr|H2L9D3|H2L9D3_ORYLA |
300.68 |
15 |
35 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101160310 PE=4 SV=1 |
|
tr|H2L9E3|H2L9E3_ORYLA |
300.68 |
15 |
35 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101160310 PE=4 SV=1 |
|
tr|G1FCF2|G1FCF2_SINCH |
284.42 |
81 |
46 |
2 |
Tropomyosin OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|W5JYT8|W5JYT8_ASTMX |
275.40 |
74 |
41 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MA79|H2MA79_ORYLA |
275.17 |
78 |
44 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101164789 PE=3 SV=1 |
|
tr|H2MCV5|H2MCV5_ORYLA |
272.04 |
62 |
37 |
2 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101172748 PE=3 SV=1 |
|
tr|W5L2L4|W5L2L4_ASTMX |
271.87 |
8 |
37 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5KS79|W5KS79_ASTMX |
269.00 |
64 |
36 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5JZC9|W5JZC9_ASTMX |
265.32 |
70 |
39 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5K3X6|W5K3X6_ASTMX |
264.76 |
10 |
24 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2L9I1|H2L9I1_ORYLA |
261.73 |
10 |
21 |
0 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|W5KTD3|W5KTD3_ASTMX |
244.75 |
57 |
30 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|V9KFU2|V9KFU2_CALMI |
242.28 |
52 |
26 |
0 |
Tropomyosin alpha-3 chain-like protein (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9KPC1|V9KPC1_CALMI |
242.28 |
51 |
26 |
0 |
Tropomyosin1-1 (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MRC9|H2MRC9_ORYLA |
241.80 |
49 |
28 |
2 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101160801 PE=3 SV=1 |
|
tr|H2LUJ8|H2LUJ8_ORYLA |
240.90 |
44 |
23 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes PE=3 SV=1 |
|
tr|W5L9M3|W5L9M3_ASTMX |
240.03 |
10 |
19 |
3 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYH13 (4 of 4) PE=4 SV=1 |
|
tr|V9K7A6|V9K7A6_CALMI |
235.45 |
10 |
19 |
1 |
Myosin, heavy chain 7, cardiac muscle, beta OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5K855|W5K855_ASTMX |
222.23 |
9 |
18 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus GN=MYH13 (2 of 4) PE=4 SV=1 |
|
tr|D2KQG2|D2KQG2_SINCH |
220.91 |
49 |
12 |
6 |
Parvalbumin 3 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|V9LBJ3|V9LBJ3_CALMI |
219.92 |
38 |
18 |
0 |
Tropomyosin alpha-1 chain-like protein (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MYK1|H2MYK1_ORYLA |
215.55 |
59 |
15 |
0 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|H2MUU9|H2MUU9_ORYLA |
214.46 |
30 |
19 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101167740 PE=3 SV=1 |
|
tr|H2MUU8|H2MUU8_ORYLA |
214.46 |
29 |
19 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101167740 PE=3 SV=1 |
|
tr|V5RFJ8|V5RFJ8_SINCH |
214.15 |
8 |
18 |
2 |
Slow skeletal muscle myosin heavy chain isoform 1 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|C6L8J0|C6L8J0_ORYLA |
205.48 |
7 |
17 |
1 |
Ventricular myosin heavy chain OS=Oryzias latipes GN=vmhc PE=2 SV=1 |
|
tr|H2MDF6|H2MDF6_ORYLA |
205.48 |
7 |
17 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=vmhc PE=4 SV=1 |
|
tr|W5KA98|W5KA98_ASTMX |
200.86 |
8 |
15 |
0 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYH13 (3 of 4) PE=4 SV=1 |
|
tr|W5L7Y3|W5L7Y3_ASTMX |
198.60 |
33 |
21 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5KAS6|W5KAS6_ASTMX |
195.33 |
38 |
20 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2L9Z5|H2L9Z5_ORYLA |
186.03 |
48 |
8 |
4 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=mylz2 PE=4 SV=1 |
|
tr|W5KKQ1|W5KKQ1_ASTMX |
184.10 |
54 |
10 |
3 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2LNN2|H2LNN2_ORYLA |
175.16 |
5 |
13 |
0 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|V9KCZ9|V9KCZ9_CALMI |
174.06 |
7 |
10 |
0 |
Slow myosin heavy chain 2 (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|G8GWA4|G8GWA4_SINCH |
168.04 |
46 |
7 |
0 |
Parvalbumin 1 OS=Siniperca chuatsi PE=2 SV=2 |
|
tr|A8QX86|A8QX86_ORYLA |
143.29 |
5 |
6 |
4 |
Collagen type I alpha 1 OS=Oryzias latipes GN=COL1 PE=2 SV=1 |
|
tr|H2MRA6|H2MRA6_ORYLA |
143.29 |
5 |
6 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=col1 PE=4 SV=1 |
|
tr|B7U3X4|B7U3X4_SINKN |
137.22 |
32 |
5 |
1 |
Myosin light chain 2 OS=Siniperca knerii GN=MCL PE=2 SV=1 |
|
tr|B9VJM4|B9VJM4_SINCH |
134.35 |
28 |
4 |
4 |
Troponin C OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|H2L8R0|H2L8R0_ORYLA |
134.35 |
28 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101171829 PE=4 SV=1 |
|
tr|H2L8Q7|H2L8Q7_ORYLA |
134.35 |
28 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101171829 PE=4 SV=1 |
|
tr|H2L8Q9|H2L8Q9_ORYLA |
134.35 |
28 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101171829 PE=4 SV=1 |
|
tr|B9VJM3|B9VJM3_SINCH |
129.26 |
44 |
6 |
3 |
Parvalbumin OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|W5LN49|W5LN49_ASTMX |
116.63 |
35 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus GN=ACTL8 PE=3 SV=1 |
|
tr|Q9PSV5|Q9PSV5_ORYLA |
116.63 |
29 |
4 |
4 |
Skeletal muscle actin (Fragment) OS=Oryzias latipes GN=OlMA1 PE=3 SV=1 |
|
tr|W5KS21|W5KS21_ASTMX |
116.63 |
14 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus GN=ACTG1 (2 of 2) PE=3 SV=1 |
|
tr|W5KBE9|W5KBE9_ASTMX |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|K4GLR4|K4GLR4_CALMI |
116.63 |
12 |
4 |
4 |
Cytoskeletal beta actin OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MSI7|H2MSI7_ORYLA |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101156021 PE=3 SV=1 |
|
tr|K4G324|K4G324_CALMI |
116.63 |
12 |
4 |
4 |
Actin, cytoplasmic 1 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|K4FTZ1|K4FTZ1_CALMI |
116.63 |
12 |
4 |
4 |
Actin, cytoplasmic 1 OS=Callorhynchus milii PE=2 SV=1 |
|
sp|P79818|ACTB_ORYLA |
116.63 |
12 |
4 |
4 |
Actin, cytoplasmic 1 OS=Oryzias latipes GN=actb PE=2 SV=2 |
|
tr|K4G4I8|K4G4I8_CALMI |
116.63 |
12 |
4 |
4 |
Actin, cytoplasmic 1 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|G7Z090|G7Z090_SINCH |
116.63 |
12 |
4 |
4 |
Beta-actin OS=Siniperca chuatsi GN=ACTb PE=3 SV=1 |
|
tr|H2LZV3|H2LZV3_ORYLA |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101168921 PE=3 SV=1 |
|
tr|W5K9V8|W5K9V8_ASTMX |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|K4GFR7|K4GFR7_CALMI |
116.63 |
12 |
4 |
4 |
Actin, cytoplasmic 1 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|K4GIQ3|K4GIQ3_CALMI |
116.63 |
12 |
4 |
4 |
Beta-actin OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5KBE8|W5KBE8_ASTMX |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5KQ47|W5KQ47_ASTMX |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|K4G587|K4G587_CALMI |
116.63 |
12 |
4 |
4 |
Actin, alpha 2, smooth muscle, aorta OS=Callorhynchus milii PE=2 SV=1 |
|
tr|Q6TKP3|Q6TKP3_SINCH |
116.63 |
12 |
4 |
4 |
Skeletal muscle alpha-actin OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|K4GDF4|K4GDF4_CALMI |
116.63 |
12 |
4 |
4 |
Actin, alpha 2 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|Q76N20|Q76N20_ORYLA |
116.63 |
12 |
4 |
4 |
Cardiac muscle actin OS=Oryzias latipes GN=OlMA1 PE=3 SV=1 |
|
tr|K4GEN0|K4GEN0_CALMI |
116.63 |
12 |
4 |
4 |
Actin, alpha 2 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5K0Q7|W5K0Q7_ASTMX |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5K7M9|W5K7M9_ASTMX |
116.63 |
12 |
4 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MU18|H2MU18_ORYLA |
116.63 |
12 |
4 |
4 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101170382 PE=3 SV=1 |
|
tr|K4FYQ2|K4FYQ2_CALMI |
116.63 |
11 |
4 |
4 |
Beta-actin OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5K1N1|W5K1N1_ASTMX |
115.42 |
11 |
5 |
3 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5LD34|W5LD34_ASTMX |
115.42 |
10 |
5 |
3 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2LHF2|H2LHF2_ORYLA |
109.74 |
6 |
9 |
8 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|W5L8R7|W5L8R7_ASTMX |
108.61 |
4 |
6 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5KKR2|W5KKR2_ASTMX |
108.23 |
46 |
5 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2M0U0|H2M0U0_ORYLA |
104.86 |
44 |
5 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101165806 PE=4 SV=1 |
|
tr|W5LNC0|W5LNC0_ASTMX |
95.18 |
29 |
4 |
0 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5LPS5|W5LPS5_ASTMX |
93.27 |
18 |
3 |
2 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYL3 PE=4 SV=1 |
|
tr|W5LNC1|W5LNC1_ASTMX |
89.82 |
23 |
4 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2MMQ8|H2MMQ8_ORYLA |
86.08 |
15 |
3 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101167500 PE=4 SV=1 |
|
tr|B7U3X3|B7U3X3_SINKN |
86.08 |
15 |
3 |
1 |
Myosin light chain 1 OS=Siniperca knerii GN=MCL PE=2 SV=1 |
|
tr|C7EP35|C7EP35_SINSC |
86.08 |
15 |
3 |
1 |
Myosin light chain 1 OS=Siniperca scherzeri PE=2 SV=1 |
|
tr|B6VCB3|B6VCB3_SINKN |
84.68 |
24 |
4 |
2 |
Myosin light chain 3 OS=Siniperca knerii PE=2 SV=1 |
|
tr|H2MBE4|H2MBE4_ORYLA |
82.41 |
5 |
2 |
2 |
Uncharacterized protein (Fragment) OS=Oryzias latipes PE=3 SV=1 |
|
tr|W5LI76|W5LI76_ASTMX |
79.33 |
8 |
2 |
2 |
Glyceraldehyde-3-phosphate dehydrogenase OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5LPK3|W5LPK3_ASTMX |
78.29 |
2 |
3 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2MXN4|H2MXN4_ORYLA |
77.13 |
6 |
2 |
2 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101175132 PE=3 SV=1 |
|
tr|H2MXN1|H2MXN1_ORYLA |
77.13 |
6 |
2 |
2 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101175132 PE=3 SV=1 |
|
tr|W5LLK3|W5LLK3_ASTMX |
77.13 |
6 |
2 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MXM8|H2MXM8_ORYLA |
77.13 |
6 |
2 |
2 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101175132 PE=3 SV=1 |
|
tr|G8GWA3|G8GWA3_SINCH |
76.44 |
10 |
2 |
1 |
Parvalbumin 2 OS=Siniperca chuatsi PE=2 SV=2 |
|
tr|H2M0U7|H2M0U7_ORYLA |
75.86 |
23 |
3 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101166553 PE=4 SV=1 |
|
tr|W5JXT6|W5JXT6_ASTMX |
75.12 |
1 |
2 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2M6N2|H2M6N2_ORYLA |
73.70 |
2 |
3 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101162163 PE=4 SV=1 |
|
tr|W5KH68|W5KH68_ASTMX |
73.70 |
20 |
3 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5K3P7|W5K3P7_ASTMX |
73.05 |
1 |
3 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2MW80|H2MW80_ORYLA |
71.85 |
19 |
2 |
2 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101161287 PE=3 SV=1 |
|
tr|W5K5Z0|W5K5Z0_ASTMX |
68.52 |
1 |
2 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|V9K7D3|V9K7D3_CALMI |
66.16 |
1 |
1 |
1 |
Collagen alpha-1(I) chain OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5K0S8|W5K0S8_ASTMX |
60.65 |
1 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5KAN3|W5KAN3_ASTMX |
59.59 |
6 |
3 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MNV5|H2MNV5_ORYLA |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101166392 PE=3 SV=1 |
|
tr|W5KDD1|W5KDD1_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|Q6XE27|Q6XE27_CONER |
59.26 |
66 |
1 |
1 |
Tubulin (Fragment) OS=Conus ermineus PE=4 SV=1 |
|
tr|Q65CK3|Q65CK3_CONTU |
59.26 |
24 |
1 |
1 |
Beta tubulin (Fragment) OS=Conus tulipa PE=3 SV=1 |
|
tr|W5KBI3|W5KBI3_ASTMX |
59.26 |
5 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus GN=TUBB2A PE=3 SV=1 |
|
tr|W5LD44|W5LD44_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus GN=TUBB (1 of 2) PE=3 SV=1 |
|
tr|W5LI88|W5LI88_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|V9KI87|V9KI87_CALMI |
59.26 |
4 |
1 |
1 |
Tubulin beta chain-like protein OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MCJ1|H2MCJ1_ORYLA |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101157969 PE=3 SV=1 |
|
tr|K4FSH1|K4FSH1_CALMI |
59.26 |
4 |
1 |
1 |
Beta1-tubulin OS=Callorhynchus milii PE=2 SV=1 |
|
tr|Q8UUK8|Q8UUK8_ORYLA |
59.26 |
4 |
1 |
1 |
TUBB protein OS=Oryzias latipes GN=TUBB PE=3 SV=1 |
|
tr|K4G4H2|K4G4H2_CALMI |
59.26 |
4 |
1 |
1 |
Beta1-tubulin OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5LKB5|W5LKB5_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus GN=TUBB (2 of 2) PE=3 SV=1 |
|
tr|H2LS01|H2LS01_ORYLA |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101164232 PE=3 SV=1 |
|
tr|W5KZP7|W5KZP7_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|K4GIQ2|K4GIQ2_CALMI |
59.26 |
4 |
1 |
1 |
Tubulin, beta 2C OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2LRN0|H2LRN0_ORYLA |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=tubb PE=3 SV=1 |
|
tr|H2MTT9|H2MTT9_ORYLA |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=TUBB4A PE=3 SV=1 |
|
tr|K4FTV1|K4FTV1_CALMI |
59.26 |
4 |
1 |
1 |
Tubulin, beta 2C OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2LEF2|H2LEF2_ORYLA |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101159143 PE=3 SV=1 |
|
tr|W5KEC5|W5KEC5_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5K7N6|W5K7N6_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus GN=TUBB3 PE=3 SV=1 |
|
tr|W5L9L2|W5L9L2_ASTMX |
59.26 |
4 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MPL0|H2MPL0_ORYLA |
55.44 |
2 |
1 |
1 |
Serotransferrin OS=Oryzias latipes GN=LOC100144362 PE=3 SV=1 |
|
tr|A8MN21|A8MN21_ORYLA |
55.44 |
2 |
1 |
1 |
Serotransferrin OS=Oryzias latipes PE=2 SV=1 |
|
sp|P79819|TRFE_ORYLA |
55.44 |
2 |
1 |
1 |
Serotransferrin OS=Oryzias latipes GN=tf PE=3 SV=1 |
|
tr|H2L7M4|H2L7M4_ORYLA |
54.51 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101168952 PE=3 SV=1 |
|
tr|H2L7M3|H2L7M3_ORYLA |
54.51 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101168952 PE=3 SV=1 |
|
tr|H2L7L7|H2L7L7_ORYLA |
54.51 |
4 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101168952 PE=3 SV=1 |
|
tr|H2LPB8|H2LPB8_ORYLA |
53.97 |
7 |
2 |
2 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101170788 PE=3 SV=1 |
|
tr|W5LSN2|W5LSN2_ASTMX |
50.91 |
4 |
1 |
0 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYL4 (2 of 3) PE=4 SV=1 |
|
tr|H2MKG7|H2MKG7_ORYLA |
48.00 |
7 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101170317 PE=4 SV=1 |
|
tr|H2L4J5|H2L4J5_ORYLA |
45.96 |
6 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101155923 PE=4 SV=1 |
|
tr|H2L4J9|H2L4J9_ORYLA |
45.96 |
5 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101155923 PE=4 SV=1 |
|
tr|B9VJM2|B9VJM2_SINCH |
45.96 |
5 |
1 |
1 |
Troponin I OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|H2L4K6|H2L4K6_ORYLA |
45.96 |
5 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101156490 PE=4 SV=1 |
|
tr|H2L4I4|H2L4I4_ORYLA |
45.96 |
5 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101155682 PE=4 SV=1 |
|
tr|H2L4I0|H2L4I0_ORYLA |
45.96 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101155425 PE=4 SV=1 |
|
tr|Q8JIP9|Q8JIP9_ORYLA |
44.31 |
3 |
1 |
1 |
Warm-temperature-acclimation-related-65 OS=Oryzias latipes GN=wap65 PE=2 SV=1 |
|
tr|H2M355|H2M355_ORYLA |
44.31 |
3 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=wap65 PE=4 SV=1 |
|
tr|W5LLP2|W5LLP2_ASTMX |
42.44 |
6 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2LBD5|H2LBD5_ORYLA |
42.44 |
6 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101164670 PE=4 SV=1 |
|
tr|W5LRW3|W5LRW3_ASTMX |
42.44 |
11 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|F5ANJ2|F5ANJ2_SINCH |
42.44 |
10 |
1 |
1 |
Ubiquitin a (Fragment) OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|V9LHV4|V9LHV4_CALMI |
42.44 |
9 |
1 |
1 |
Polyubiquitin-C-like protein (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5LI91|W5LI91_ASTMX |
42.44 |
9 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|Q2KKX3|Q2KKX3_SINCH |
42.44 |
8 |
1 |
1 |
Ubiquitin (Fragment) OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|H2LX76|H2LX76_ORYLA |
42.44 |
7 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101169681 PE=4 SV=1 |
|
tr|E2I6H0|E2I6H0_SINCH |
42.44 |
7 |
1 |
1 |
Ubiquitin OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|K4G4C1|K4G4C1_CALMI |
42.44 |
7 |
1 |
1 |
Ribosomal protein L40-like isoform 1 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2L6M8|H2L6M8_ORYLA |
42.44 |
4 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101167305 PE=4 SV=1 |
|
tr|W5LLW5|W5LLW5_ASTMX |
42.44 |
2 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|K4FTX6|K4FTX6_CALMI |
42.44 |
2 |
1 |
1 |
Polyubiquitin-like protein OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2M983|H2M983_ORYLA |
42.44 |
1 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101172100 PE=4 SV=1 |
|
tr|W5KHS4|W5KHS4_ASTMX |
38.98 |
10 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYL2 (2 of 3) PE=4 SV=1 |
|
tr|H2MMN3|H2MMN3_ORYLA |
38.98 |
8 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101162219 PE=4 SV=1 |
|
tr|H2MMN4|H2MMN4_ORYLA |
38.98 |
8 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101162219 PE=4 SV=1 |
|
tr|H2LZ88|H2LZ88_ORYLA |
38.98 |
7 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=MYL2 (1 of 2) PE=4 SV=1 |
|
tr|W5L038|W5L038_ASTMX |
38.42 |
0 |
1 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|K4FV77|K4FV77_CALMI |
35.11 |
6 |
1 |
1 |
MLC1f/3f OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9KS09|V9KS09_CALMI |
35.11 |
6 |
1 |
1 |
Myosin light chain 1/3, skeletal muscle isoform isoform 3f OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9LGV1|V9LGV1_CALMI |
35.11 |
6 |
1 |
1 |
Myosin light polypeptide 6B OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9LFC7|V9LFC7_CALMI |
35.11 |
6 |
1 |
1 |
Myosin, light chain 6, alkali, smooth muscle and non-muscle OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9L5Z0|V9L5Z0_CALMI |
35.11 |
5 |
1 |
1 |
Myosin light chain 1/3, skeletal muscle isoform isoform 3f (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9L470|V9L470_CALMI |
35.11 |
5 |
1 |
1 |
Myosin, light chain 4, alkali OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9KMS0|V9KMS0_CALMI |
35.11 |
5 |
1 |
1 |
Myosin, light chain 1, alkali OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MT93|H2MT93_ORYLA |
35.11 |
5 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101157706 PE=4 SV=1 |
|
tr|H2MTE5|H2MTE5_ORYLA |
35.11 |
5 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101157250 PE=4 SV=1 |
|
tr|W5KSS8|W5KSS8_ASTMX |
35.11 |
4 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2M7W2|H2M7W2_ORYLA |
30.87 |
9 |
1 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101174831 PE=4 SV=1 |
|
tr|H2LK91|H2LK91_ORYLA |
30.63 |
0 |
0 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=col2a1a PE=4 SV=1 |
|
tr|G1UH59|G1UH59_ORYLA |
30.63 |
0 |
0 |
0 |
Type II collagen A isoform 2 OS=Oryzias latipes GN=col2a1a PE=2 SV=1 |
|
tr|H2LK90|H2LK90_ORYLA |
30.63 |
0 |
0 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=col2a1a PE=4 SV=1 |
|
tr|G1UH58|G1UH58_ORYLA |
30.63 |
0 |
0 |
0 |
Type II collagen A isoform 1 OS=Oryzias latipes GN=col2a1a PE=2 SV=1 |
|
tr|W5LKA3|W5LKA3_ASTMX |
30.03 |
1 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=4 SV=1 |
|
total 194 proteins |
Table 3: List of detected proteins in spray-dried C. striatus samples from a combination of whole sample and fractions analysis
|
Accession |
Max -10lgP |
(%) Max coverage |
Max # peptides |
Max # unique |
Description |
|
tr|Q05K05|Q05K05_ORYLA |
187.31 |
33 |
108 |
2 |
Fast skeletal myosin heavy chain isoform mMYH-11 OS=Oryzias latipes PE=4 SV=1 |
|
tr|H2LNX8|H2LNX8_ORYLA |
185.20 |
61 |
106 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=MYH13 (4 of 11) PE=4 SV=1 |
|
tr|Q05K07|Q05K07_ORYLA |
184.06 |
31 |
101 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-7 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q6SNT2|Q6SNT2_SINCH |
184.03 |
29 |
96 |
6 |
Fast skeletal muscle myosin heavy chain OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|G0YU48|G0YU48_SINCH |
184.03 |
29 |
96 |
6 |
Fast skeletal muscle myosin heavy chain isoform 1 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|G0YU49|G0YU49_SINCH |
184.03 |
29 |
96 |
6 |
Fast skeletal muscle myosin heavy chain isoform 2 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|G0YU50|G0YU50_SINCH |
184.03 |
29 |
96 |
6 |
Fast skeletal muscle myosin heavy chain isoform 3 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|Q05K09|Q05K09_ORYLA |
183.87 |
33 |
106 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-2 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q05K08|Q05K08_ORYLA |
182.40 |
29 |
95 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-3 OS=Oryzias latipes PE=4 SV=1 |
|
tr|H2LPD7|H2LPD7_ORYLA |
182.07 |
29 |
93 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=MYH13 (6 of 11) PE=4 SV=1 |
|
tr|Q05K06|Q05K06_ORYLA |
181.98 |
32 |
101 |
0 |
Fast skeletal myosin heavy chain mMYH-9 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q05K11|Q05K11_ORYLA |
181.58 |
32 |
100 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-5 OS=Oryzias latipes PE=4 SV=1 |
|
tr|H2N1T3|H2N1T3_ORYLA |
181.41 |
32 |
92 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=MYH13 (11 of 11) PE=4 SV=1 |
|
tr|H2LPK6|H2LPK6_ORYLA |
179.79 |
30 |
97 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=MYH13 (7 of 11) PE=4 SV=1 |
|
tr|Q05K12|Q05K12_ORYLA |
179.40 |
30 |
94 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-6 OS=Oryzias latipes PE=4 SV=1 |
|
tr|Q05K10|Q05K10_ORYLA |
178.52 |
29 |
92 |
0 |
Fast skeletal myosin heavy chain isoform mMYH-1 OS=Oryzias latipes PE=4 SV=1 |
|
tr|C4TIP2|C4TIP2_ORYLA |
178.34 |
26 |
81 |
0 |
Myosin heavy chain larval type 1 OS=Oryzias latipes GN=mMYHL1 PE=2 SV=1 |
|
tr|C4TIP3|C4TIP3_ORYLA |
178.24 |
27 |
81 |
0 |
Myosin heavy chain larval type 2 OS=Oryzias latipes GN=mMYHL2 PE=2 SV=1 |
|
tr|C4TIP1|C4TIP1_ORYLA |
177.92 |
26 |
90 |
0 |
Myosin heavy chain embryonic type 1 OS=Oryzias latipes GN=mMYHemb1 PE=2 SV=1 |
|
tr|H2L9D3|H2L9D3_ORYLA |
176.38 |
24 |
84 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101160310 PE=4 SV=1 |
|
tr|H2L9E3|H2L9E3_ORYLA |
176.38 |
24 |
84 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101160310 PE=4 SV=1 |
|
tr|W5L2L4|W5L2L4_ASTMX |
173.01 |
13 |
82 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|V9K7A6|V9K7A6_CALMI |
161.02 |
18 |
66 |
6 |
Myosin, heavy chain 7, cardiac muscle, beta OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5K3X6|W5K3X6_ASTMX |
158.95 |
19 |
57 |
5 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2L9I1|H2L9I1_ORYLA |
157.53 |
19 |
52 |
0 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|W5L9M3|W5L9M3_ASTMX |
148.03 |
15 |
44 |
4 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYH13 (4 of 4) PE=4 SV=1 |
|
tr|W5L0R5|W5L0R5_ASTMX |
146.58 |
10 |
39 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|V5RFJ8|V5RFJ8_SINCH |
146.10 |
13 |
44 |
3 |
Slow skeletal muscle myosin heavy chain isoform 1 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|G1FCF2|G1FCF2_SINCH |
144.79 |
75 |
48 |
3 |
Tropomyosin OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|C6L8J0|C6L8J0_ORYLA |
141.71 |
11 |
37 |
0 |
Ventricular myosin heavy chain OS=Oryzias latipes GN=vmhc PE=2 SV=1 |
|
tr|H2MDF6|H2MDF6_ORYLA |
141.71 |
11 |
37 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=vmhc PE=4 SV=1 |
|
tr|H2MA79|H2MA79_ORYLA |
141.21 |
69 |
45 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101164789 PE=3 SV=1 |
|
tr|W5JYT8|W5JYT8_ASTMX |
139.36 |
71 |
41 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5KA98|W5KA98_ASTMX |
136.71 |
11 |
30 |
0 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYH13 (3 of 4) PE=4 SV=1 |
|
tr|H2MDC8|H2MDC8_ORYLA |
135.02 |
12 |
37 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=mmyhc1 PE=4 SV=1 |
|
tr|W5K2C5|W5K2C5_ASTMX |
134.18 |
9 |
27 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2MCV5|H2MCV5_ORYLA |
132.50 |
68 |
36 |
8 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101172748 PE=3 SV=1 |
|
tr|W5KS79|W5KS79_ASTMX |
131.30 |
55 |
32 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MYK1|H2MYK1_ORYLA |
130.16 |
79 |
25 |
0 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|W5K855|W5K855_ASTMX |
129.27 |
12 |
29 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus GN=MYH13 (2 of 4) PE=4 SV=1 |
|
tr|V9KFU2|V9KFU2_CALMI |
128.41 |
58 |
30 |
1 |
Tropomyosin alpha-3 chain-like protein (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|V9KPC1|V9KPC1_CALMI |
128.41 |
57 |
30 |
1 |
Tropomyosin1-1 (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2LNN2|H2LNN2_ORYLA |
126.92 |
12 |
37 |
0 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|H2MLX5|H2MLX5_ORYLA |
126.82 |
9 |
30 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101160127 PE=4 SV=1 |
|
tr|V9KCZ9|V9KCZ9_CALMI |
126.60 |
19 |
34 |
0 |
Slow myosin heavy chain 2 (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MRC9|H2MRC9_ORYLA |
125.16 |
53 |
30 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101160801 PE=3 SV=1 |
|
tr|V9LBJ3|V9LBJ3_CALMI |
111.64 |
40 |
18 |
0 |
Tropomyosin alpha-1 chain-like protein (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2L9Z5|H2L9Z5_ORYLA |
109.49 |
70 |
14 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=mylz2 PE=4 SV=1 |
|
tr|V9KBQ3|V9KBQ3_CALMI |
109.08 |
10 |
21 |
0 |
Myosin-7B-like protein (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5L7Y3|W5L7Y3_ASTMX |
105.31 |
31 |
20 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5KKQ1|W5KKQ1_ASTMX |
102.15 |
53 |
13 |
5 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5KBE9|W5KBE9_ASTMX |
102.08 |
25 |
14 |
14 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|K4GDF4|K4GDF4_CALMI |
102.08 |
23 |
14 |
14 |
Actin, alpha 2 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5KBE8|W5KBE8_ASTMX |
102.08 |
23 |
14 |
14 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|Q76N20|Q76N20_ORYLA |
102.08 |
23 |
14 |
14 |
Cardiac muscle actin OS=Oryzias latipes GN=OlMA1 PE=3 SV=1 |
|
tr|W5KQ47|W5KQ47_ASTMX |
102.08 |
23 |
14 |
14 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|K4GEN0|K4GEN0_CALMI |
102.08 |
23 |
14 |
14 |
Actin, alpha 2 OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5K0Q7|W5K0Q7_ASTMX |
102.08 |
23 |
14 |
14 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
sp|Q98972|ACTS_ORYLA |
102.08 |
23 |
14 |
14 |
Actin, alpha skeletal muscle OS=Oryzias latipes GN=acta1 PE=2 SV=1 |
|
tr|Q6TKP3|Q6TKP3_SINCH |
102.08 |
23 |
14 |
14 |
Skeletal muscle alpha-actin OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|W5K7M9|W5K7M9_ASTMX |
102.08 |
23 |
14 |
14 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|K4G587|K4G587_CALMI |
102.08 |
23 |
14 |
14 |
Actin, alpha 2, smooth muscle, aorta OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MU18|H2MU18_ORYLA |
102.08 |
23 |
14 |
14 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101170382 PE=3 SV=1 |
|
tr|D2KQG2|D2KQG2_SINCH |
99.67 |
49 |
10 |
4 |
Parvalbumin 3 OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|W5L0H6|W5L0H6_ASTMX |
97.77 |
35 |
15 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5KT30|W5KT30_ASTMX |
96.00 |
52 |
11 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|B7U3X4|B7U3X4_SINKN |
94.15 |
66 |
11 |
2 |
Myosin light chain 2 OS=Siniperca knerii GN=MCL PE=2 SV=1 |
|
tr|W5K709|W5K709_ASTMX |
92.83 |
44 |
10 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|G8GWA4|G8GWA4_SINCH |
87.40 |
58 |
8 |
1 |
Parvalbumin 1 OS=Siniperca chuatsi PE=2 SV=2 |
|
tr|H2MRA6|H2MRA6_ORYLA |
76.88 |
7 |
10 |
7 |
Uncharacterized protein OS=Oryzias latipes GN=col1 PE=4 SV=1 |
|
tr|A8QX86|A8QX86_ORYLA |
76.88 |
7 |
10 |
7 |
Collagen type I alpha 1 OS=Oryzias latipes GN=COL1 PE=2 SV=1 |
|
tr|H2LHF2|H2LHF2_ORYLA |
69.38 |
7 |
12 |
11 |
Uncharacterized protein OS=Oryzias latipes PE=4 SV=1 |
|
tr|B9VJM4|B9VJM4_SINCH |
66.94 |
24 |
4 |
4 |
Troponin C OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|H2L8R0|H2L8R0_ORYLA |
66.94 |
24 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101171829 PE=4 SV=1 |
|
tr|H2L8Q7|H2L8Q7_ORYLA |
66.94 |
24 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101171829 PE=4 SV=1 |
|
tr|H2L8Q9|H2L8Q9_ORYLA |
66.94 |
24 |
4 |
4 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101171829 PE=4 SV=1 |
|
tr|W5L8R7|W5L8R7_ASTMX |
64.09 |
4 |
9 |
4 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5LNC1|W5LNC1_ASTMX |
61.90 |
24 |
5 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2MXN4|H2MXN4_ORYLA |
61.74 |
6 |
3 |
3 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101175132 PE=3 SV=1 |
|
tr|H2MXN1|H2MXN1_ORYLA |
61.74 |
6 |
3 |
3 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101175132 PE=3 SV=1 |
|
tr|W5LLK3|W5LLK3_ASTMX |
61.74 |
6 |
3 |
3 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MXM8|H2MXM8_ORYLA |
61.74 |
6 |
3 |
3 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101175132 PE=3 SV=1 |
|
tr|W5KKR2|W5KKR2_ASTMX |
60.48 |
47 |
6 |
0 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5KHS4|W5KHS4_ASTMX |
59.79 |
26 |
3 |
0 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYL2 (2 of 3) PE=4 SV=1 |
|
tr|W5LI76|W5LI76_ASTMX |
59.21 |
8 |
3 |
3 |
Glyceraldehyde-3-phosphate dehydrogenase OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2M0U0|H2M0U0_ORYLA |
58.82 |
44 |
5 |
0 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101165806 PE=4 SV=1 |
|
tr|B6VCB3|B6VCB3_SINKN |
57.86 |
27 |
5 |
2 |
Myosin light chain 3 OS=Siniperca knerii PE=2 SV=1 |
|
tr|W5LPS5|W5LPS5_ASTMX |
54.69 |
18 |
3 |
3 |
Uncharacterized protein OS=Astyanax mexicanus GN=MYL3 PE=4 SV=1 |
|
tr|W5JXT6|W5JXT6_ASTMX |
54.46 |
3 |
6 |
3 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|B9VJM3|B9VJM3_SINCH |
54.34 |
41 |
5 |
1 |
Parvalbumin OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|W5LNC0|W5LNC0_ASTMX |
53.48 |
29 |
4 |
0 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|W5LPK3|W5LPK3_ASTMX |
53.14 |
2 |
4 |
2 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2MMQ8|H2MMQ8_ORYLA |
52.72 |
19 |
4 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101167500 PE=4 SV=1 |
|
tr|B7U3X3|B7U3X3_SINKN |
52.72 |
19 |
4 |
1 |
Myosin light chain 1 OS=Siniperca knerii GN=MCL PE=2 SV=1 |
|
tr|C7EP35|C7EP35_SINSC |
52.72 |
19 |
4 |
1 |
Myosin light chain 1 OS=Siniperca scherzeri PE=2 SV=1 |
|
tr|V9KUX9|V9KUX9_CALMI |
51.39 |
15 |
3 |
3 |
Triosephosphate isomerase OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MB61|H2MB61_ORYLA |
51.39 |
14 |
3 |
3 |
Triosephosphate isomerase (Fragment) OS=Oryzias latipes GN=tpi-b PE=3 SV=1 |
|
tr|V9L1I9|V9L1I9_CALMI |
49.53 |
12 |
3 |
3 |
Troponin T type 3 (Skeletal, fast) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5K1N1|W5K1N1_ASTMX |
47.89 |
4 |
2 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5LD34|W5LD34_ASTMX |
47.89 |
4 |
2 |
2 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|H2MBE4|H2MBE4_ORYLA |
46.20 |
5 |
2 |
2 |
Uncharacterized protein (Fragment) OS=Oryzias latipes PE=3 SV=1 |
|
tr|V9LAW6|V9LAW6_CALMI |
45.28 |
18 |
2 |
1 |
Myosin light chain 2V (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2M0U7|H2M0U7_ORYLA |
44.36 |
24 |
4 |
0 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101166553 PE=4 SV=1 |
|
tr|W5KH68|W5KH68_ASTMX |
42.82 |
20 |
4 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2LAV5|H2LAV5_ORYLA |
41.98 |
21 |
4 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101165624 PE=4 SV=1 |
|
tr|W5K3M6|W5K3M6_ASTMX |
41.88 |
4 |
3 |
3 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5K3P7|W5K3P7_ASTMX |
41.65 |
1 |
3 |
1 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2M6N2|H2M6N2_ORYLA |
39.68 |
2 |
5 |
1 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101162163 PE=4 SV=1 |
|
tr|G8GWA3|G8GWA3_SINCH |
38.87 |
10 |
2 |
1 |
Parvalbumin 2 OS=Siniperca chuatsi PE=2 SV=2 |
|
tr|V9K8V8|V9K8V8_CALMI |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2LN45|H2LN45_ORYLA |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Oryzias latipes GN=LOC101171864 PE=3 SV=1 |
|
tr|H2LN49|H2LN49_ORYLA |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Oryzias latipes GN=LOC101171864 PE=3 SV=1 |
|
tr|W5LFK4|W5LFK4_ASTMX |
37.44 |
5 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus GN=ATP2A1 (1 of 2) PE=4 SV=1 |
|
tr|H2M9N1|H2M9N1_ORYLA |
37.44 |
3 |
1 |
1 |
Calcium-transporting ATPase OS=Oryzias latipes GN=ATP2A1 (2 of 2) PE=3 SV=1 |
|
tr|V9KH80|V9KH80_CALMI |
37.44 |
2 |
1 |
1 |
Sarcoplasmic/endoplasmic reticulum calcium ATPase 2-like protein (Fragment) OS=Callorhynchus milii PE=2 SV=1 |
|
tr|W5LM60|W5LM60_ASTMX |
37.44 |
2 |
1 |
1 |
Uncharacterized protein OS=Astyanax mexicanus GN=ATP2A1 (2 of 2) PE=4 SV=1 |
|
tr|W5KSD8|W5KSD8_ASTMX |
37.44 |
2 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|G1EJ40|G1EJ40_SINCH |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|H2M858|H2M858_ORYLA |
37.44 |
2 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101171024 PE=4 SV=1 |
|
tr|H2M854|H2M854_ORYLA |
37.44 |
2 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101171024 PE=4 SV=1 |
|
tr|W5L8A3|W5L8A3_ASTMX |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|V9KCE0|V9KCE0_CALMI |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2M9M8|H2M9M8_ORYLA |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Oryzias latipes GN=ATP2A1 (2 of 2) PE=3 SV=1 |
|
tr|H2MMF8|H2MMF8_ORYLA |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Oryzias latipes GN=LOC101160819 PE=3 SV=1 |
|
tr|H2M9N3|H2M9N3_ORYLA |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Oryzias latipes GN=ATP2A1 (2 of 2) PE=3 SV=1 |
|
tr|H2MMF9|H2MMF9_ORYLA |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Oryzias latipes GN=LOC101160819 PE=3 SV=1 |
|
tr|W5LA91|W5LA91_ASTMX |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|V9K9A3|V9K9A3_CALMI |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2MMF7|H2MMF7_ORYLA |
37.44 |
2 |
1 |
1 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101160819 PE=4 SV=1 |
|
tr|W5KQZ1|W5KQZ1_ASTMX |
37.44 |
2 |
1 |
1 |
Calcium-transporting ATPase OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|V9K7D3|V9K7D3_CALMI |
36.23 |
2 |
3 |
1 |
Collagen alpha-1(I) chain OS=Callorhynchus milii PE=2 SV=1 |
|
tr|H2L470|H2L470_ORYLA |
33.57 |
4 |
2 |
2 |
Uncharacterized protein OS=Oryzias latipes GN=LOC101166239 PE=3 SV=1 |
|
tr|C7ASM1|C7ASM1_SINCH |
33.57 |
4 |
2 |
2 |
Muscle-type creatine kinase OS=Siniperca chuatsi PE=2 SV=1 |
|
tr|H2L804|H2L804_ORYLA |
33.57 |
4 |
2 |
2 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101163677 PE=3 SV=1 |
|
tr|W5KP12|W5KP12_ASTMX |
33.57 |
4 |
2 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=3 SV=1 |
|
tr|W5K5Z0|W5K5Z0_ASTMX |
32.92 |
2 |
3 |
2 |
Uncharacterized protein OS=Astyanax mexicanus PE=4 SV=1 |
|
tr|H2LPB8|H2LPB8_ORYLA |
30.14 |
7 |
2 |
2 |
Uncharacterized protein (Fragment) OS=Oryzias latipes GN=LOC101170788 PE=3 SV=1 |
|
total 137 proteins |
Collagen
Two types of collagen have been detected in both C. striatus water extract samples, which are type I and type II collagen. Collagen is a natural substrate for cellular attachment, growth and differentiation, and promotes cellular proliferation and differentiation. As described by Sonnemann and Bement (2011), a large amount of collagen and other extracellular matrix (ECM) proteins are secreted by the fibroblasts and myofibroblasts into the wound area, resulting in the basis of the so-called granulation tissue. Then the myofibroblasts contract the granulation tissue and the cells associated with it, closing the wound slowly and at the same time aligning the collagen fibres that composed the ECM. Many publications have shown the effect of collagen involved in the wound healing process (Kwan et al., 2011; Ruszczak, 2003). A recent study has shown that the use of concentrated collagen hydrogel favoured cell proliferation and also protected fibroblasts against apoptosis during the treatment of a chronic skin wound (Helary et al., 2012). Thus, collagen detected in the C. striatus sample may be another key component that assists in the wound healing process.
Uncharacterised Protein
Uncharacterised proteins consisted of more than 50% of the total proteins detected. The functions of such proteins are still unknown as the information is unavailable. However, with the high coverage detected using the PEAKS studio database matching, we are certain that the proteins exist in the sample. The uncharacterised proteins are designated as proteins of unknown functions due to no detectable homology to proteins of known functions at both the sequence and structure level (Lubec et al., 2005). Nadzirin and Firdaus-Raih (2012) has mentioned that the functions of uncharacterised proteins have not been characterised from assays following the structural analyses. The lack of information on a protein’s characterised function restricts the further exploitation of these proteins. Nevertheless, there is still a possibility that one or more of the uncharacterised proteins detected in our samples are relevant to the wound healing property of C. striatus.
Overall, the proteome profiling using the high sensitivity LTQ-Orbitrap Velos Pro mass spectrometer has allowed us to explore the protein content in the C. striatus water extracts. It is evident that the extracts, which resemble local delicacy haruan soup, contain a considerable amount and variety of proteins. The high amount of uncharacterised proteins detected also show that the proteome database for C. striatus is not yet complete and requires more extensive work on the matter. Freeze-dry technique was shown to retain slightly higher amounts and variety of proteins in the water extracts than the spray-dry technique. Other parts of C. striatus, such the mucus layer and flesh, are currently being similarly studied. Overall, the current data has given us the insight on the proteins available in the C. striatus and a more detailed work has to be conducted to determine the proteins that are involved in accelerating the wound healing process.
Conflict of Interest
The authors agree that there is no conflict of interest.
Authors’ contribution
Soon Hong Kwan conductdt the experiment under the supervision of Mohd Nazri Ismail. Saringat Baie and Nornisah Mohammed contributed with their previous experiences for solving some of the difficulties encountered during the entire project.
Acknowledgement
This study was supported by APEX DE 2012 grants (1002/PFARMASI/910332) (1002/PDOPING/910335). The studentship of Kwan, Soon Hong is supported by the USM Fellowship.
References