Journal of Animal Health and Production
Research Article
Isolation and Detection of Four Major Virulence Genes in O157:H7 and Non-O157 E. Coli from Beef at Yogyakarta Special Province, Indonesia
Alvita Indraswari1, Aris Haryanto2, I Wayan Suardana3, Dyah Ayu Widiasih4*
1Graduate School of Veterinary Science, Faculty of Veterinary Medicine, Universitas Gadjah Mada, Jl. Fauna No. 2 Karangmalang, Yogyakarta 55281, Indonesia; 2Department of Biochemistry, Faculty of Veterinary Medicine, Universitas Gadjah Mada, Jl. Fauna No. 2 Karangmalang, Yogyakarta 55281, Indonesia; 3Department of Veterinary Public Health, Faculty of Veterinary Science, University of Udayana, Jl. P.B. Sudirman Denpasar, Bali, Indonesia; 4Department of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Gadjah Mada, Jl. Fauna No. 2 Karangmalang, Yogyakarta 55281, Indonesia.
Abstract | Food products of animal origin, especially beef, are at risk of Escherichia coli (E. coli) contamination during slaughtering and/or processing that poses serious threat to consumers. E. coli contamination affects food quality because the pathogen contains toxins that can cause disease and even death. This study aimed to detect virulence genes in E. coli O157:H7 and non-O157 isolates of beef sold in the traditional markets of the Yogyakarta Special Province, Indonesia, using the multiplex polymerase chain reaction (multiplex PCR). A total of 98 raw beef samples were collected aseptically from different markets of study area and cultured on various selective media followed by the analysis by multiplex PCR amplification. The results exhibited that out of 98 beef samples, 83 (84,69%) were positive for E. coli. The main virulence genes detected were eae, fliC, rfbE, and stx2A. All of these virulence genes of E. coli O157:H7 were detected in all beef samples. The 28 isolates were identified as non-O157 E. coli that exhibited the eae gene, however they were not exhibited the rfbE and fliC genes. This study concluded the presence of significant contamination of virulent E. coli strains in beef meat sold in the Yogyakarta Special Province, Indonesia. Moreover, multiplex PCR assay was shown to be effective for the detection of E. coli O157:H7 and non-O157 strains from beef and may be applied for other food matrices as well.
Keywords | Beef, Escherichia coli O157, Indonesia, Multiplex PCR, Virulence
Received | May 17, 2021; Accepted | May 21, 2021; Published | September 25, 2021
*Correspondence | Dyah Ayu Widiasih, Department of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Gadjah Mada, Jl. Fauna No. 2 Karangmalang, Yogyakarta 55281, Indonesia; Email: [email protected]
Citation | Indraswari A, Haryanto A, Suardana IW, Widiasih DA (2021). Isolation and detection of four major virulence genes in O157:H7 and non-O157 E. coli from beef at yogyakarta special province, indonesia. J. Anim. Health Prod. 9(4): 371-379.
DOI | http://dx.doi.org/10.17582/journal.jahp/2021/9.4.371.379
ISSN | 2308-2801
Copyright © 2021 Widiasih et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Escherichia coli O157:H7 is a subset of enterohemorrhagic E. coli (EHEC) that can cause diarrhea in human. Escherichia coli O157:H7 differ from normal flora found in the intestine and are categorized into specific group based on virulence factors, such as exotoxin (Fernandez, 2008). The EHEC produces Shiga-like toxin, it is also called Shiga toxin (stx)-producing E. coli (STEC). Stx consists of two subgroups, namely stx1 and stx2. Other virulence factors found in EHEC are eae (intimin), rfbE (O157 antigen), and fliC (flagellar antigen). EHEC lives in animal intestines and can be transferred to humans through contaminated food (CDC, 2016). Cattle are known as the main reservoir of E. coli O157:H7, but the strain also found in other animals such as sheep, goats, cats, chickens, gulls (Nataro and Kaper, 1998) and pigs (Goma et al., 2019; Tseng et al., 2014). According to Widiasih et al. (2003), the greatest excretion of E. coli O157:H7 was observed in naturally infected cattle suffering from food poisoning by STEC. Most infected animals do not indicate any clinical symptoms, although young animals may suffer enteric disease caused by members of some non-O157 serogroups (Spickler, 2016). Escherichia coli O157: H7 is one of the most important foodborne infectious agents. Transmission of this bacterium to humans mainly occurs through consumption of undercooked meat, unpasteurized animal products, and feces contaminated water as well as human-to-human contact (Fernandez, 2008). According to some previous reports (Hastuti et al., 2015; Rinca et al., 2016; Suardana et al., 2010; Baehaqi et al., 2015) in Bali, Indonesia the occurrence of E. coli O157:H7 in beef, cattle feces, and human feces indicates the high zoonotic potential of the serotype; thus, further studies on virus epidemiology and virulence factors were strongly suggested.
The high level of similarity between E. coli O157: H7 isolates found in humans and cattle indicates a high zoonotic potential (Suardana et al., 2015). In the United States, E. coli O157:H7 was the second leading cause of foodborne diseases after Salmonella sp. in 1997; in this year, 3,260 cases of disease and eight deaths were attributed to this pathogen (CDC, 1997). E. coli O157:H7 can contaminate every product of animal origin and the environment (Kiranmayi et al., 2010; WHO, 2020). In Indonesia, E. coli O157:H7 was observed in beef, fresh and pasteurized milk, water samples, the hands of slaughterhouse and dairy workers, and beef traders (Sartika et al., 2005; Cho et al., 2020; Emara et al., 2016). Generally, the multiplex polymerase chain reaction (multiplex PCR) method can rapidly and reliably identify foodborne agents, such as E. coli O157:H7 and other diarrheagenic E. coli, in a sustainable manner (Nguyen et al., 2016; Ibrahim et al., 2016; Taha and Yassin, 2019). Multiplex PCR is known to detect six main types of virulence factors found in E. coli O157:H7, namely, the fliC, stx1, stx2, eae, rfbE, and hlyA genes (Bai et al., 2010). The technique has also been used to detect the Z3276, a unique target gene of E. coli O157:H7, (Li et al., 2017).
While going through the literature, it was observed that studies are lacking on the foodborne pathogens, especially E. coli O157:H7, in beef that sold in traditional markets in the Yogyakarta Special Province, Indonesia. Thus this study aimed the rapid detection of four major virulence genes, eae, fliC, rfbE, and stx2A in E. coli O157:H7 isolated from beef that was being sold in the traditional markets in the Yogyakarta Special Province, Indonesia, using the multiplex PCR. The present research is expected to support the government’s efforts to improve food safety and provide a sense of security when consuming food products from animals, especially beef. Moreover, this survey results can assist researchers in detecting E. coli O157:H7 rapidly and effectively and highlight the need to process beef properly to prevent foodborne disease.
Materials and Methods
Ethical approval
This study used beef samples and bacterial isolates thus no ethical approval was needed.
Collection and preparation of beef samples
Beef samples were obtained from traditional markets in Yogyakarta City, including the Beringharjo, Kranggan, Ngasem, Demangan, Lempuyangan, and Serangan markets. Beef samples (approximately 100 g) were collected aseptically from sellers and packed and labeled in sterile plastic bags. The samples were transported in ice boxes containing ice gel to the Veterinary Public Health Laboratory, Faculty of Veterinary Medicine, Universitas Gadjah Mada. The number of beef samples to be tested was determined according to the formula of Martin et al. (1987). Based on an estimated prevalence of 5.62% (Suardana et al., 2007) with a desired error of 5%, the number of samples required for testing with a confidence level of 95% were a minimum of 85 samples. Thus, a total of 98 samples were collected and analyzed in this study.
Isolation and identification of Escherichia coli
The bacterial isolation procedure in this study referred to Suardana et al. (2007). Each sample was aseptically removed and placed in a sterile plastic bag containing a peptone buffer solution (Conda CAT: 1403.00) at a ratio of 1:9. The plastic was then placed in a stomacher for 60 sec. Peptone buffer rinsing solution was used to isolate E. coli. Up to 100 µl of this rinsing solution was cultured on eosin methylene blue agar (EMBA) medium (Oxoid CM0069) via the spreading method. The culture was incubated at 37 °C for 24 h. E. coli colonies were identified by their metallic green sheen and a black spot in the middle of a colony. Colonies positive for E. coli in the EMBA medium were then stored in liquid brain heart infusion (BHI) medium (Conda CAT: 1400.00) added with glycerol for further analysis.
Positive isolates on the EMBA media were then cultured in selective sorbitol MacConkey agar (SMAC) medium (Oxoid CM 0813). After incubation at 37 °C for 24 h, sorbitol-negative E. coli colonies were identified via their clear and colorless appearance and then stored in liquid
Table 1: Primers used to detect four major virulence genes in E. coli O157:H7 and non-O157 E. coli
| Primer | Primer sequence (5’-3’) | Target genes (product size) | Reference |
| FLICH7 F | GCG CTG TCG AGT TCT ATC GAG C |
fliC (625 bp) |
|
| FLICH7 R | CAA CGG TGA CTT TAT CGC CAT TCC | ||
| rfbE-F | TCT TTC CTC TGC GGT CCT A |
rfbE (317 bp) |
|
| rfbE-R | CAG GTG AAG GTG GAA TGG T | ||
| stx2A-F | CGA GGG CTT GAT GTC TAT CAG |
stx2A (553 bp) |
|
| stx2A-R | TCA GTA TAA CGG CCA CAG TCC | ||
| eae-F | CAT TAT GGA ACG GCA GAG GT |
eae (375 bp) |
|
| eae-R | ACG GAT ATC GAA GCC ATT TG |
Table 2: Biochemical tests results of Escherichia coli isolates.
| No. | Sample code | Tests | Gram staining | ||||||||
| Glucose | Lactose | Mannitol | Maltose | Sucrose | Sulfur | Indole | Motility | Simmons citrate | |||
| 1 | D1 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
|
2 |
D2 | + | + | + | + | - | - | + | + | - | Negative rod |
| 3 | KR9 | + | + | + | + | + | - | + | + | - | Short Negative rod |
| 4 | B6 | + | + | + | + | + | - | + | + | - | Short Negative rod |
| 5 | B4 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
| 6 | B3 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
| 7 | B2 | + | + | + | + | - | - | + | + | - | Negative coccobacilli |
| 8 | B7-2 | + | + | + | + | + | - | + | + | - | Short Negative rod |
| 9 | B12-2 | + | + | + | + | + | - | + | + | - | Short negative rod |
| 10 | B8-3 | + | + | + | + | + | - | - | + | - | Negative coccobacilli |
| 11 | B14-3 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
|
12 |
B3-3 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
| 13 | B12-3 | + | + | + | + | + | - | - | + | - | Negative coccobacilli |
| 14 | B16-2 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
| 15 | KR8-2 | + | + | + | + | - | - | + | + | - | Negative coccobacilli |
| 16 | KR1-2 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
| 17 | KR3-2 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
| 18 | KR1-3 | + | + | + | + | + | - | + | + | - | Negative coccobacilli |
|
19 |
N2 | + | + | + | + | + | - | + | + | - | Negative rod |
| 20 | D1-2 | + | + | + | + | + | - | + | + | - | Negative rod |
| 21 | D2-2 | + | + | + | + | + | - | + | + | - | Negative rod |
| 22 | D4-2 | + | + | + | + | + | - | + | + | - | Negative rod |
| 23 | D1-3 | + | + | + | + | + | - | + | + | - | Negative rod |
| 24 | D4-3 | + | + | + | + | + | - | + | + | - | Negative rod |
| 25 | N1-2 | + | + | + | + | + | - | + | + | - | Negative rod |
| 26 | N2-2 | + | + | + | + | + | - | + | + | - | Negative rod |
| 27 | N2-3 | + | + | + | + | + | - | + | + | - | Negative rod |
| 28 | L2-2 | + | + | + | + | + | - | + | + | - |
Negative rod |
+: positive; -: negative
BHI medium added with glycerol for further analysis. Gram staining to differentiate bacteria as Gram-negative or -positive and biochemical tests, including glucose, lactose, mannitol, maltose, sucrose, sulfur, indole, motility, and Simmons citrate tests, were subsequently carried out.
DNA extraction
DNA extraction was carried out using a Presto™ Mini gDNA Bacteria Kit (Geneaid GBB300) according to the manufacturer’s instructions. First, bacterial cells were centrifuged at 14,000 rpm for 1 min. Then, the supernatant was discarded, and the cell pellet was resuspended in 180 µl of GT buffer and vortexed. The cell pellet was mixed with 20 µl of proteinase K and incubated at 60 ºC for 10 min with inversion every 3 min. Next, the solution was added with 200 µl of GB buffer, mixed for 10 sec, and then incubated at 70 °C for 10 min with inversion every 3 min.
The required elution buffer was heated to 70 °C and then added with 200 µl of absolute ethanol and mixed. This solution was transferred to the GD column and centrifuged at 14,000 rpm for 2 min. The collection tube was discarded, and the solution was added with 400 µl of W1 buffer and centrifuged at 14,000 rpm for 30 sec. The supernatant was discarded once more, and the cell pellet was added with 600 µl of wash buffer and centrifuged at 14,000 rpm for 30 sec. This process was repeated one more time. Finally, the column was transferred to a microcentrifuge tube, added with 100 μl of pre-heated elution buffer, allowed to stand for 3 min, and then centrifuged at 14,000 rpm for 30 sec. The suspension was used for target DNA in molecular analysis.
Multiplex PCR standardization for E. coli O157:H7 detection
Multiplex PCR standardization was performed to detect different E. coli species. Each reaction mixture contained 2 µl of the DNA templates, 1 µl each of the forward and reverse primers, and 12.5 µl of MyTaq™ HS Red Mix (Cat. No.: BIO-25048). UltraPure™ ddH2O (Cat No.: 10977015) was added to achieve a total volume of 25 µl. The reaction mixture was vortexed until a homogeneous solution was obtained and amplified using a personal MasterCycler. Primers derived from the eae, fliC, rfbE, and stx2A genes were obtained from IDT™ (Table 1) to detect E. coli O157:H7. Amplification was carried out on a gradient PCR machine at annealing temperatures of 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, and 61 °C simultaneously. PCR amplification was carried out as follows: initial denaturation at 94 °C for 5 min; 40 cycles of denaturation at 94 °C for 45 sec for, annealing at 50 °C to 61 °C for 30 sec, and extension at 72 °C for 45 sec; final extension at 72 °C for 10 min. Approximately 5 µl of the amplification result was analyzed by gel electrophoresis (1.5% agarose gel stained with SYBR® Safe) in 1× TBE solution. The gel was visualized with a UV transilluminator and photographed by a digital camera.
Detection of non-O157 E. coli
The target genes selected for non-O157 E. coli detection were the eae, fliC, and rfbE genes. Primers derived from the eae, fliC, and rfbE genes were obtained from IDT™ (Table 1) to detect non-O157 E. coli. The reaction mixture contained 2 µl of the DNA template, 1 µl each of the forward and reverse primers, and 12.5 µl MyTaq™ HS Red Mix (Cat No.: BIO-25048). UltraPure™ DNase/RNase-free distilled water (Cat No.: 10977015) was added to achieve a total volume of 25 µl. PCR amplification was carried out as follows: initial denaturation at 94 °C for 5 min; 40 cycles of denaturation at 94 °C for 45 sec, annealing at 54 °C for 30 sec, and extension at 72 °C for 45 sec; final extension at 72 °C for 10 min. Approximately 5 µl of the amplification result was analyzed by gel electrophoresis (1.5% agarose gel stained with SYBR® Safe) in a 1× TBE solution. The gel was visualized with a UV transilluminator and photographed using a digital camera.
Statistical analysis
Data obtained from the study were analyzed descriptively using the Microsoft Word and presented as tables and figures.
Results
Occurrence of Escherichia coli in meat samples
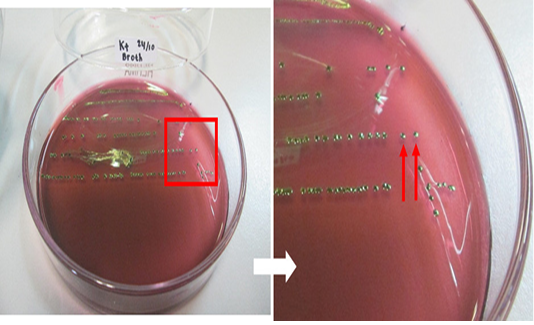
The results of the microbiological investigation of E. coli are shown in Figure 1. Colonies showing a green metallic sheen with a black spot in the middle were identified on EMBA medium as E. coli. In this study, out of 98 beef samples 83 (84.69%) were positive for E. coli. Subsequently, 28 samples (D1, D2, KR9, B6, B9, B4, B3, B2, B7-2, B12-2, B8-3, B14-3, B3-3, B12-3, B16-2, KR8-2, KR1-2, KR3-2, KR1-3, N2, D1-2, D2-2, D4-2, D1-3, D4-3, N1-2, N2-2, N2-3, and L2-2) were tested as a presumption to indicate E. coli O157 on SMAC medium. E. coli colonies that do not ferment sorbitol were characterized by a clear or colorless appearance, as shown in Figure 2. The positive control isolate of E. coli O157:H7 showed the same results when cultured on EMBA and SMAC media. E. coli is characterized by its pink colonies and short rod-shaped structure during Gram staining. E. coli also showed positive results in the glucose, lactose, mannitol, maltose, sucrose, and indole tests but negative results in the Simmons citrate test. The results of biochemical tests proved the presence of E. coli. The results of biochemical tests on 28 isolates were presumed to be E. coli O157 listed in Table 2.
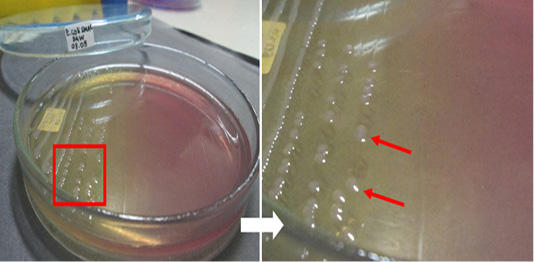
Figure 2: Escherichia coli O157-positive control isolate culture on SMAC medium. The red arrow indicates the colonies.
Multiplex PCR standardization for detection the eae, fliC, rfbE, and stx2A genes of E. coli O157:H7
The multiplex PCR protocol was carried out as described under materials and methods. The results of multiplex PCR optimization using a positive control isolate of E. coli O157:H7 with two target genes are shown in Figure 3. The multiplex PCR was carried out using two target genes simultaneously, namely, fliC and rfbE, with product sizes of 625 and 317 bp, respectively. The DNA bands of the two target genes was found according to their product size at different annealing temperatures. Well numbers 1 to 12 in the Figure-3 showed the resultant DNA bands in annealing temperature used from 50 to 61 °C.
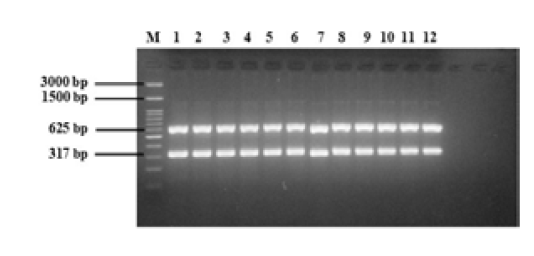
Figure 3: Multiplex PCR optimization of E. coli O157:H7 with two target genes (fliC, 625 bp; rfbE, 317 bp). M = 100 bp DNA marker, 1 to 12 = annealing temperatures 50 to 61°C.
The next round of optimization was carried out using four target genes, namely, eae, fliC, rfbE, and stx2A, which have product sizes of 375, 625, 317, and 553 bp, respectively. The results of multiplex PCR optimization with four target genes are shown in Figure 4. The DNA bands of the four target genes was found according to their product size at different annealing temperatures. Well numbers 1 to 12 in the figure-4 showed the resultant DNA bands in annealing temperature used from 50 to 61 °C.
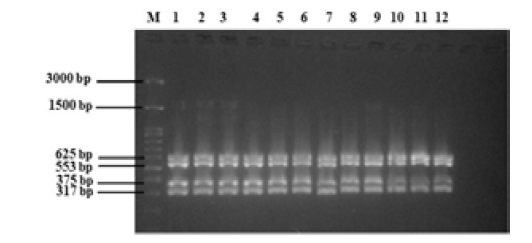
Figure 4: Multiplex PCR standardization E. coli O157:H7 with four target genes (eae, 375 bp; fliC, 625 bp; rfbE, 317 bp; stx2A, 553 bp). M = 100 bp DNA marker, 1 to 12 = annealing temperatures 50 to 61°C.
Detection the eae, fliC, and rfbE genes of non-O157 E. coli
The standardized protocol was used to screen the field samples. The results of eae gene detection from 28 E. coli samples from beef are shown in Figures 5 and 6. The fliC and rfbE genes were also detected in these 28 samples, however no isolate had rfbE and fliC genes (data not shown). In this study, all tested E. coli isolates were positive for eae gene (100%). The eae gene produced DNA bands with a size of 375 bp. Our results indicated that the PCR method may differentially detected all three virulence genes, eae, fliC, and rfbE, in non-O157 serotypes.
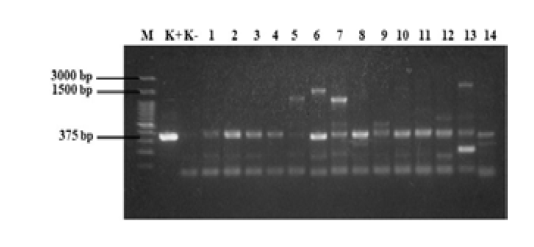
Figure 5: PCR results of samples. M = 100 bp DNA marker, K+ = positive control, K- = negative control, 1 = D1, 2 = D2, 3 = KR9, 4 = B6, 5 =B4, 6 =B3, 7 =B2, 8 = B7-2, 9 =B12-2, 10= B8-3, 11 = B14-3, 12 = B3-3, 13 = B12-3, and 14 = B16-2. Annealing temperature, 53 °C.
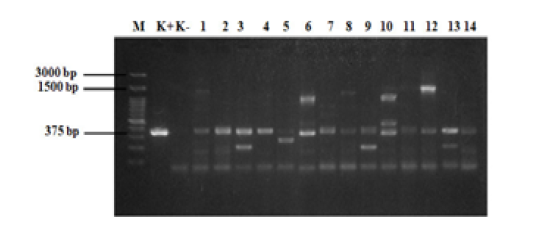
Figure 6: PCR results of samples. M = 100 bp DNA marker, K+ = positive control, K- = negative control, 1 =KR8-2, 2 = KR1-2, 3 = KR3-2, 4 = KR1-3, 5 = N2, 6 = D1-2, 7 = D2-2, 8 = D4-2, 9= D1-3, 10 = D4-3, 11 = N1-2, 12 = N2-2, 13 = N2-3, and 14 = L2-2. Annealing temperature 53 °C.
Discussion
E. coli O157: H7 is one of the strains of EHEC which was first identified as a human pathogen in 1982. At that time there were cases of hemorrhagic colitis in fast-food restaurants in Oregon and Michigan, United States (Fernandez, 2008; Besser et al., 1999). Since then, E. coli O157: H7 has been identified to be responsible for various outbreaks and sporadic cases around the world.
The results of bacteriological investigation of the beef samples revealed the 84.6% (83/98) meat samples had E. coli. On SMAC medium, 33.7% (28/83) of the samples presumed to be E. coli O157. Identification of 28 (33.7 %) E. coli isolates out of the 83 collected samples was confirmed by biochemical tests using standard bacteriological procedures. The 28 isolates were visualized as colonies with a green metallic sheen and a black spot in the middle of the colonies on EMBA medium, while bright pink or colorless colonies on SMAC medium. These findings indicated a presumptive identification of E. coli that was confirmed using standard bacteriological procedures.
The results of this study were higher than the findings of different authors. Zulfakar et al. (2017) reported that 53.3% (24/45) of the beef samples obtained from wet markets are contaminated by E. coli. Soepranianondo et al. (2019) showed that 32.5% (13/40) of the meat samples obtained from a slaughterhouse in East Java Province, Indonesia, were contaminated by E. coli. Carcasses are mainly contaminated with the microbes during slaughtering and processing in abattoirs. E. coli is commonly found in all environments, but some serotypes may be extremely harmful to humans. The prevalence of E. coli O157 and other than O serotype was recorded 30% and 41% in a study used 820 samples of beef carcasses obtained from abattoirs in Alberta, Canada (Manage et al., 2019). The study further proposed that this contamination in beef by STEC (most common cause of foodborne diseases) may occur during evisceration and handling procedures.
Multiplex PCR standardization was performed on positive control isolate of E. coli O157:H7. The results of the amplification of fliC and rfbE genes using PCR (Figure 3) revealed that the control isolate of E. coli O157:H7 had fliC and rfbE genes. In previous studies, the optimum annealing temperatures for the fliC and rfbE genes were found to be 58 °C (Al-Ajmi et al., 2006) and 57 °C (Suria et al., 2013), respectively. However, in the present study, we used gradient PCR, the optimum annealing temperatures for subsequent PCR included 52, 53, 54, and 55 °C (well 3 to 6 of Figure 3). Our findings confirmed the identity of isolates as E. coli O157:H7 and proved that fliC and rfbE genes were present in E. coli O157:H7 as reported by Bai et al. (2010). rfbE and fliC are genes encoding for the O157 somatic and H7 flagellar antigens respectively (Wang et al., 2002). Two sets of primers specifically designed to amplify the fliC and rfbE genes (O157 antigen) were observed only in the E. coli O157:H7 strain. The primer pair for the E. coli H7 fliC gene was highly specific for the H7 flagella antigen. Thus, only E. coli strains with alternative O antigen serotypes (e.g., O114:H7) could show positive results when analyzed via the multiplex PCR method (Al-Ajmi et al., 2006; Suria et al., 2013) using these primers. According to Botkin et al. (2012) the multiplex PCR method permitted differentiation of EPEC (enteropathogenic E. coli), STEC, and EHEC strains from other pathogenic E. coli, therefore the assay becomes an additional tool for rapid diagnostic of these organisms.
The next cycle of multiplex PCR standardization was carried out by using four target genes. The target genes used were eae, fliC, rfbE, and stx2A, which have product sizes of 375, 625, 317, and 553 bp, respectively. The results of the amplification of eae, fliC, rfbE, and stx2A genes using PCR (Figure 4) revealed that the control isolate of E. coli O157:H7 showed positive result for the presence of these four genes. In previous studies, the optimal annealing temperatures for the eae and stx2A genes were 65 °C (Bai et al., 2010) and 61 °C (Park et al., 2006), respectively. However, in the present study, gradient PCR was employed, and the optimum annealing temperatures for subsequent PCR used 52 and 53 °C (wells 3 and 4 of Figure 4). The main virulence factor possessed by E. coli O157: H7 is related to its ability to produce Shiga-toxin (stx). Stx consists of two major groups, namely stx1 and stx2, each group is divided into several subtypes, namely stx1a, 1c, and 1d; stx2a to stx2e. Not all E. coli O157: H7 isolates can produce both types of toxins. Some specific strains can produce both (stx1 and stx2), but some strains only produce one type of toxin, stx1 or stx2 (Spickler, 2016). Strains that carry only the stx2 gene are known to have higher pathogenicity. Intimine is produced by the eae gene, is an attaching and effacing gene (gene species-specific) for E. coli firmly attached to the intestine (ehxA gene) localizing high toxin levels that cause hemolytic disease (Manage et al., 2019). Strains that have the eae and stx2 genes are known to be more pathogenic to humans and the stx2 gene, in particular, is an important virulence factor associated with the causes of the hemolytic uremic syndrome and hemorrhagic colitis in humans (Kesaya et al., 2011). E. coli O157: H7 is most commonly found in ruminants. Widiasih et al. (2003) has proven that naturally infected cattle were known to be able to secrete the stx1 and stx2 genes through feces over a long time, therefore it has the potential to induce disease in humans.
A singleplex PCR detecting eae gene was performed on E. coli isolated from beef. The results of the amplification of eae gene using PCR (Figure 5 and 6) revealed that all E. coli isolates showed positive result for the presence of eae gene, thus confirming their identity as pathogenic E. coli. The eae gene codes an adhesion protein called intimin, which attaches and removes microvilli on the surface of host cell enterocytes. The protein also plays an essential role on the pathogenicity of the EPEC pathotype of the bacteria (Xu et al., 2016). In order to detection the serotypes of pathogenic E. coli, multiplex PCR was used for detection of rfbE and fliC genes. No isolate had rfbE and fliC genes (data not shown). In this study, the eae gene produced DNA bands with a size of 375 bp and all positive samples had the eae gene. Figure 5 shows that 14 samples tested positive for the eae gene but samples B4, B3, B2, B12-2, B3-3, and B12-3 (wells 5, 6, 7, 9, 12, and 13) also produced products that were not target genes. Figure 6 shows that 14 samples tested positive for the eae gene but samples D1-2, D4-2, D4-3, and N2-2 (wells 6, 8, 10, and 12) also produced DNA bands that were not the target gene. Bai et al. (2010), reported the 65 °C as the optimum annealing temperature for the eae gene. The optimum annealing temperature determined in this study was 53 °C because several DNA bands besides from the target gene were produced at 52 °C (data not shown). Further research needs to be done using a higher annealing temperature to determine the DNA bands produced.
Suardana et al. (2011), reported that 95% of clinical and non-clinical human, beef, cow feces, and chicken feces samples were positive for eae gene. The presence of the eae gene indicates that the E. coli isolates have an adhesion molecule called intimin. E. coli can colonize in the intestine, induce attaching and effacing (AE) lesions with the help of intimin, and cause a cytopathic effect on intestinal epithelial cells. The ability to induce AE lesions is encoded by pathogenicity islands, the locus of enterocyte effacement, which has genes involved in the formation of AE lesions. Intimin is a virulence factor of EPEC and EHEC E. coli strains (Al-Ajmi et al., 2006); thus, further research is necessary to ensure the type of pathotype.
Conclusion
E. coli O157: H7 is a foodborne disease agent that is dangerous based on its virulence factors. Our results demonstrated that beef sold in the market of Yogyakarta Special Province, Indonesia is potentially contaminated by pathogenic E. coli. Thus vigorous attention and monitoring of consumers, veterinarians, and relevant stakeholders are needed to prevent foodborne diseases. Moreover, multiplex PCR was recognized as a valid and rapid method for detecting virulence genes in E. coli. This multiplex assay was shown to be effective for the detection of E. coli O157:H7 and screening for non-O157 strains in beef, and may be applied for other food matrices as well.
Authors contributions
AI, AH, IWS, and DAW designed the study. AI conducted the field surveys and analyzed samples. AH contributed to the PCR testing. IWS and DAW analyzed the data and coordinated the study. All authors read and approved the final manuscript.
Acknowledgements
The authors express their gratitude to The Directorate of Research and Community Service, Directorate General of Research and Development Strengthening, The Ministry of Research, Technology and Higher Education, Republic of Indonesia for funding this research under the 2019 PMDSU research grant scheme (Contract No. 2952/UN1.DITLIT/DIT-LIT/LT/2019).
Conflict of Interest
The authors declare that they have no conflict of interests.
References