Advances in Animal and Veterinary Sciences
Research Article
Some Studies on Phenotypic and Genotypic Characters of Small Colony Variants Staphylococcus aureus Isolated from Dairy Cows Infected with Mastitis in Egypt
Shereen S. El Mohandes1, Inas M. Gamal2, Hala A. Abou-Zeina3, Mohamed K. Elbayoumy3*
1Mastitis and neonatal diseases department. Animal reproduction research institute (ARRI), Giza, Egypt; 2Immunology and Immunopharmacology unit. Animal reproduction research institute (ARRI), Giza, Egypt; 3Parasitology and Animal Diseases Research dept., Veterinary Division, National Research Centre (NRC), Giza, Egypt.
Abstract | In veterinary medicine, the small colony variants (SCVs) S.aureus was ignored, but recently the interest in its analysis has been increased because it causes chronic infections with difficulty in laboratory isolation and identification. Therefore, this study aimed to investigate the phenotypic and genotypic characteristics of S. aureus SCVs and their mastitis prevalence in dairy cows, as well as the protein profile discrepancy between parent strains of S.aureus and S.aureus SCVs. A total number of 130 milk samples were collected and examined from commercial Friesian-Holstein dairy herds in Egypt. Of the examined milk samples chronic subclinical mastitic milk samples constituted 58.46%, while clinical mastitis milk samples were 9.23%. S.aureus SCVs were only isolated from chronic subclinical samples with percentage of 23.68%. In order to distinguish between S.aureus parent strains and S.aureus SCVs, Phenotypic characters such as, growth rate, colony size, Mannitol fermentation, hemolysis, rabbit plasma coagulation and catalase test were examined. In addition the isolated Staphylococcus species were investigated using Polymerase Chain Reaction (PCR) technique to approve the isolation of SCVs S. aureus. In addition protein profile between parent strains of S.aureus and SCVs have been tested and evaluated. The results showed the significance of SCVs S.aureus in herd health, as a result of difficulty of its diagnosis, the percentage of SCVs infection may be increased, so more attention should be paid for the investigation of observed colony morphologies. Isolates of SCVs S.aureus should be subjected to antimicrobial susceptibility testing with a protocol for treatment as they showed higher resistance against many antibiotics when compared with parent strains of S.aureus; this study suggests that Ciprofloxacin, Enrofloxacin and rifampin can be used for treatment of SCVs S.aureus.
Keywords | Subclinical, Mastitis, S.aureus, SCVs S.aureus, PCR & protein profile.
Received | December 23, 2020; Accepted | January 01, 2021; Published | March 01, 2021
*Correspondence | Mohamed K Elbayoumy, Parasitology and Animal Diseases Research dept., Veterinary Division, National Research Centre (NRC), Giza, Egypt; Email: [email protected]
Citation | Mohandes SSE, Gamal IM, Abou-Zeina HA, Elbayoumy MK (2021). Some studies on phenotypic and genotypic characters of small colony variants staphylococcus aureus isolated from dairy cows infected with mastitis in egypt. Adv. Anim. Vet. Sci. 9(5): 637-647.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.5.637.647
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Elbayoumy is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Staphylococcus species is a major causative agent of bovine mastitis. Some Staphylococcus aureus (S.aureus) strains are able to switch to an alternative isolates that are having specific phenotypic characteristics such as slow growth, small colonies and decreased hemolysis, therefore these bacterial variants termed the small colony variants (SCVs) of S.aureus (Tuasikal et al., 2012; Zhu et al., 2016). SCVs originate by mutations in metabolic genes, causing in appearance of auxotrophic bacterial subpopulations and these isolates allowing their persistence under stressful circumstances (Bui et al., 2015). The phenomenon of the generation of a slower-growing phenotype causing in smaller colonies on solid agar media is not limited to members of the Staphylococcus genus. As described for staphylococcal SCVs, the first reports on Salmonella enteric serovarTyphi“Eberthella typhosa” and other Salmonella serovars, Serratia marcescens, Vibrio cholerae, and Shigella species were published at the beginning of the 20th century (Swingle, 1935).
In human medicine, S. aureus SCVs have gained more attention, as they have been recovered from patients with endocarditis, pneumonia, soft tissue infections, osteomyelitis, and severe bacteremia, but in veterinary medicine it have been underestimated and overlooked (McNamara & Proctor, 2000). Recently in veterinary field some researches proved that SCVs were isolated from milk samples collected from dairy cows with history of chronic mastitis (Atalla et al., 2008; Zhu et al., 2016). Persistent S. aureus infections in animals may be related to SCVs that can hide inside host cells, persist longer and at greater numbers within non-phagocytes than do their parentages and , modulate host defenses which may explain antimicrobial therapy and vaccine failures (Atalla et al., 2010). Therefore the prevalence of antibiotic-resistant bacteria cause a global healthcare issue with dramatically limited therapeutic choices (Brandis et al., 2017).
(Atshan et al., 2015) stated that Two-dimensional gel electrophoresis (2DGE) is a well-known traditional tool that is broadly useful for protein separation and quantitation. This method was originally applied on biological fluids to identify potential biomarkers and study model organisms such as Escherichia coli and Bacillus subtilis. Doan et al. (2013) used SDS- PAGE for typing methicillin resistant S.aureus strains (MRSA) got from the various regions of patients in intensive care unit (ICU). The results of the previous study in MRSA strains by SDS-PAGE and earlier studies clearly designated that valuable epidemiological information can be verified with electrophoretic methods.
This study was designed for examination and isolation of SCVs S. aureus from dairy cows with bovine mastitis infection. The prevalence and phenotypic characters were investigated using Polymerase Chain Reaction (PCR) technique as a sensitive accurate technique in diagnosis of direct detection of low concentrations of bacteria or bacterial products to approve isolation of SCVs S. aureus. Antimicrobial resistance testing against it has also been observed because a little is known about the effect of antibiotics on SCVs S.aureus. Furthermore, one of the goals of the current investigation was to compare between the protein profile of the SCVs S.aureus and parent S.aureus isolates using Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS-PAGE) in a trial to understand and characterize possible protein markers that could indicate and identify possible differences in protein expression among those isolates which seems important in combating of mastitis caused by SCVs S.aureus.
Materials and Methods
Collection of milk samples
A total of 130 milk samples were collected from cows raised in private dairy farms located in different governorates of Egypt (Alexandria -Bani suif- El Fayoum and EL Gharbia). After clinical examination of udder, California mastitis test (CMT) was applied for detection of subclinical mastitis and milk samples were aseptically collected as previously described by Amer et al. (2018).
Bacteriological culture and identification of small colony variants S.aureus
Collected milk samples were examined as formerly designated by Quinn et al. (2011) samples were cultured on Mannitol salt agar media and blood agar media (complemented with 5% sheep blood) for isolation of Staphylococcus spp. Investigation of phenotypic characters as Mannitol fermentation and detection of hemolysis were tested. Coagulase test using rabbit plasma and catalase test using H2O2 were also established.
Antimicrobial Susceptibility Testing
Using disk diffusion technique on Mueller Hinton agar, isolated strains of parent S.aureus and SCVs S.aureus were subjected to 13 commercially available antibiotics discs [Ciprofloxacin; CIP (5μg), Enrofloxacin; ENR (5μg), amoxicillin-clavulanic acid; AMC (30μg), amoxicillin; AMX (25 μg), penicillin; P (10 U), tetracycline; TE (30μg), florofenicol: FFC (30μg), gentamycin; CN (10μg), streptomycin; S (10μg) and cefoquinom; CEQ (30μg), Rifampin; RD (5μg), Kanamycin (30μg) and trimethoprim 1.25 μg plus sulfamethoxazole 23.75 μg (SXT) and the inhibition zones were recorded according to the “National Committee for Clinical Laboratory Standards”(Shryock et al., 2008).
Preparation of genomic DNA and PCR procedures (genotypic identification)
DNA was extracted from bacterial colonies using Nucleic Acid Extraction kit (Vivantis (GF-1) according to the manufacturer’s recommendations. Genomic DNA concentrations were determined spectrophotometrically by (Nanodrop Micro volume spectrophotometer Quawell Q9000CM), and the DNA samples were stored at −20°C until PCR was accomplished.
PCR was performed for detection of the probable SCVs; they were recognized as S.aureus SCVs using (16srDNA, nucA, and nuc) primers demonstrated in Table (1) as previously recorded by Zhu, et al. (2016), to a total volume of 25 μl using a master mix (Solis BioDyne 5x), specific primers at a concentration of 0.25 mM for each primer in Table (1), and the DNA template as recommended by the
Table 1: Primer sequence according to Zhu, et al. 2016
| Sequence | Target gene | Tm | Amplicon |
|
Forward primer: 5'-GGCGTTGCTCCGTCAGGCTT-3' Reverse primer: 5'-CGCTGGCGGCGTGCCTAAT-3' |
16SrDNA | 54°C | 375 bp |
|
Forward primer: 5'-CGCTTGCTATGATTGTGGTAGCC-3' Reverse primer: 5'-TTCGGTTTCACCGTTTCTGGCG-3' |
nucA | 239 bp | |
|
Forward primer: 5'-TCGTCAAGGCTTGGCTAAAGTTGC-3' Reverse primer: 5'-TCAGCGTTGTCTTCGCTCCAAA-3' |
nuc | 126 bp |
Table 2: Incidence of Mastitis in examined milk samples
| No. of milk samples | Type of milk samples | |||||
| Normal milk | Mastitic milk | |||||
| Subclinical mastitic milk | Clinical mastitic milk | |||||
| No. | % | No. | % | No. | % | |
| (130) | 42 | 32.31 | 76 | 58.46 | 12 | 9.23 |
Table 3: Phenotypic characters of parent S.aureus and SCVs S.aureus.
|
Type of detected S.aureus |
Growth rates | Colony size | Mannitol fermentation | Hemolysis | Coagulation of rabbit plasma | Catalase test |
|
Parent S.aureus |
Rapid (18-24) hours | Large | Positive | Mostly hemolytic | Positive | Positive |
|
SCVs S.aureus |
Very slow (up to 48 - 72 hours) | Very small | Negative | Mostly non hemolytic | Slow or Negative | Positive |
PCR kit producer, all of the PCR parameters were consisted to the following thermo-cycling: 1 cycle of 95°C for 5 min, 34 cycles of 95°C for 30 sec, 54°C for 30 sec, 72°C for 1 min, 1 cycle of 72°C for 5 min and, 4°C for ∞ In a T100™ Thermal Cycler Bio-Rad, Laboratories, Inc.. Then the amplification product was analyzed by electrophoresis in 2% agarose gel.
Interpretation of the PCR products
Using agarose gel electrophoresis, PCR products were separated by a 100 bp DNA ladder (100bp DNA Ladder h3 RTU, Gene Direx) as a molecular indicator on 2% agarose (ABgene), containing non-mutagenic fluorescent reagent (Novel Juice, Gene Direx) and visualized using ultraviolet light trans-illuminator.
Protein Profile using Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS-PAGE):
1-Preparation of parent S. aureus and SCVs S. aureus strains for PAGE: Bacterial isolates grew in Brain Heart Infusion (BHI) broth, were collected by centrifugation at 10,000g for 10 minutes, washed with physiological saline, diluted by physiological saline to 1.5 ml/v. Cell suspension was sonicated for 3 minutes (at level 4), then centrifuged at 11,000g for 3 minutes. Supernatant obtained was taken and stored at 0-4 ºC for further analysis according to Tuasikal et al. (2012).
2-Polyacrylamide gel electrophoresis: Analysis of proteins using Coomassie blue staining was achieved by typical protocols of (Laemmli, 1970). Selected SCVs S. aureus strains where submitted to SDS-PAGE and their protein patterns were compared with a database of protein fingerprints derived from parent S. aureus.
3-Computer-aided analysis of the gels: Using a sharp JX-330 flat-bed scanner, images of the gels were captured and image examination of the protein profiles was completed using Amersham Pharmacia Biotech Image master 2-D Elite software.
RESULTS
As presented in Table (2), inspection of a total number of 130 milk samples exposed that 12 (9.23%) of milk samples were collected from cows suffered from clinical mastitis with signs of inflammation (hotness, redness, swelling and pain of the udder). Using CMT for apparently normal cows, 42 (32.31%) of examined milk samples were CMT negative, not infected with subclinical mastitis, while 76 (58.46%) were collected from cows suffered from subclinical mastitis and reacted positively with CMT.
After cultivation of milk samples on Mannitol salt agar and blood agar media, coagulase and catalase tests were applied on Staphylococcus isolates. The phenotypic characters
Table 4: Identification of SCVs S.aureus by PCR technique.
|
Staphylococcus spp. (55) |
Phenotypic characters | PCR | ||
| NO (Total) | % | NO (Total) | % | |
|
Parent S.aureus (35) |
35 | 26.92 | 35 | 26.92 |
|
SCVs S.aureus (20) |
20 | 15.38 | 18 | 13.85 |
Table 5: Differentiation and identification of different Staphylococcus spp. isolated from the examined milk samples using both phenotypic and genotypic methods
|
Type of Staphylococcus spp. |
Type of milk samples |
Total milk samples (130) |
||||||
|
Normal milk (42) |
Mastitiic milk (88) |
|||||||
| Subclinical (76) | Clinical (12) | |||||||
| No. | % | No. | % | No. | % | No. | % | |
|
Parent S.aureus |
1 | 2.38 | 27 | 35.53 | 7 | 58.33 | 35 |
26.92 |
|
SCVs S.aureus |
0 | 0 | 18 | 23.68 | 0 | 0 | 18 | 13.85 |
|
Other Staphylococcus spp. (CNS) |
4 | 9.52 | 15 | 19.74 | 2 | 16.67 | 21 |
16.15 |
*Percentage was calculated according to total No. of each group.
Table 6: Antibiotic sensitivity against parent S.aureus and SCVs S.aureus.
| Antibiotic discs |
Parent S.aureus (35 isolates) |
SCVs S.aureus (18 isolates) |
||||||||||
| Sensitive | Intermediate | Resistance | Sensitive | Intermediate | Resistance | |||||||
| No | % | No | % | No | % | No | % | No | % | No | % | |
| Ciprofloxacin | 28 | 80 | 5 | 14.29 | 2 | 5.71 | 13 | 72.22 | 2 | 11.11 | 3 | 16.66 |
| Enrofloxacin | 30 | 85.71 | 2 | 5.71 | 3 | 8.58 | 12 | 66.67 | 2 | 11.11 | 4 | 22.22 |
|
Amoxicillin- clavulanic acid |
24 | 67.57 | 7 | 20 | 4 | 11.43 | 8 | 44.44 | 3 | 16.66 | 7 | 38.90 |
| Amoxicillin | 0 | 0 | 3 | 8.57 | 32 | 91.43 | 0 | 0 | 1 | 5.56 | 17 |
94.44 |
| Penicillin | 2 | 5.71 | 7 | 20 | 26 | 74.29 | 0 | 0 | 5 | 27.78 | 13 | 72.22 |
| Tetracycline | 3 | 8.57 | 4 | 11.43 | 28 | 80 | 1 | 5.56 | 3 | 16.66 | 14 | 77.78 |
| Florofenicol | 25 | 71.42 | 5 | 14.29 | 5 | 14.29 | 3 | 16.66 | 5 | 27.78 | 10 | 55.56 |
| Gentamycin | 22 | 62.86 | 9 | 25.71 | 4 | 11.43 | 2 | 11.11 | 6 | 33.33 | 10 | 55.56 |
| Streptomycin | 0 | 0 | 8 | 22.86 | 27 | 77.14 | 0 | 0 | 0 | 0 | 18 | 100 |
| Kanamycin | 3 | 8.57 | 9 | 25.71 | 23 | 65.72 | 0 | 0 | 4 | 22.22 | 14 | 77.78 |
| Cefoquinom | 31 | 88.58 | 2 | 5.71 | 2 | 5.71 | 3 | 16.66 | 4 | 22.22 | 11 | 61.11 |
| Rifampin | 28 | 80 | 5 | 14.29 | 2 | 5.71 | 16 | 88.90 | 1 | 5.55 | 1 | 5.55 |
| SXT | 19 | 54.29 | 7 | 20 | 9 | 25.71 | 2 | 11.11 | 5 | 27.78 | 11 |
61.11 |
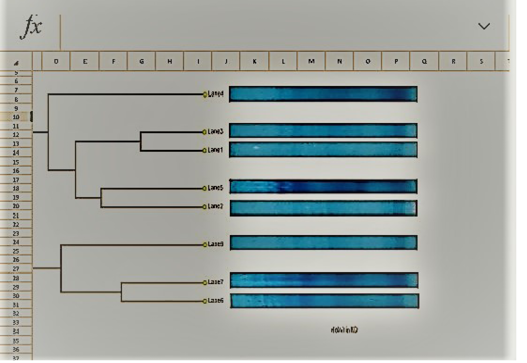
Table 7: Dendrogram analysis of the isolated parent S.aureus versus SCVs S.aureus
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 | Lane 7 | Lane 8 | |
| Lane 1 | 1 | 0.43 | 0.71 | 0.4 | 0.27 | 0 | 0 | 0.27 |
| Lane 2 | 0.43 | 1 | 0.57 | 0.53 | 0.13 | 0 | 0 | 0 |
| Lane 3 | 0.71 | 0.57 | 1 | 0.4 | 0.27 | 0 | 0 | 0.13 |
| Lane 4 | 0.4 | 0.53 | 0.4 | 1 | 0.13 | 0.13 | 0.25 | 0.12 |
| Lane 5 | 0.27 | 0.13 | 0.27 | 0.13 | 1 | 0.53 | 0.40 | 0.38 |
| Lane 6 | 0 | 0 | 0 | 0.13 | 0.53 | 1 | 0.63 | 0.38 |
| Lane 7 | 0 | 0 | 0 | 0.25 | 0.40 | 0.63 | 1 | 0.63 |
| Lane 8 | 0.27 | 0 | 0.13 | 0.12 | 0.38 | 0.38 | 0.63 | 1 |
of parent S.aureus and SCV S.aureus were explained in Table (3) and illustrated in Figures 1, 2 and 3 which demonstrated the differences in growth rate, colony size, Mannitol fermentation, hemolysis, plasma coagulation and catalase test results between parent or wild strains of S.aureus and SCVs.

Figure 3: Differences between parent S.aureus (Positive) and SCVs S.aureus (Negative) in coagulase test
Table (3) showed that parent strains of S.aureus grew faster with larger yellow colonies and they were able to ferment Mannitol and coagulate rabbit plasma. On blood agar media parent strains had hemolytic colonies. On the other hand, SCVs slowly grew up to 72 hrs forming small, colorless colonies, and fail to ferment Mannitol. SCV S.aureus strains grew on blood agar as non-hemolytic, non-pigmented pinpoint colonies and their biochemical tests were non-reactive.
Diagnosis of SCVs S.aureus is very difficult and consumed long time for its identification therefore molecular identification of parent S.aureus and SCVs S.aureus was shown in Table (4) comparing to phenotypic characters and biochemical identification of Staphylococcus isolates. Table (4) and Figure 4, 5 and 6 recorded that parent S.aureus strains and SCVs S.aureus represented 26.92% and 15.38%, respectively of isolates by phenotypic examination while SCVs S.aureus was decreased to 13.84% of isolates by molecular identification.
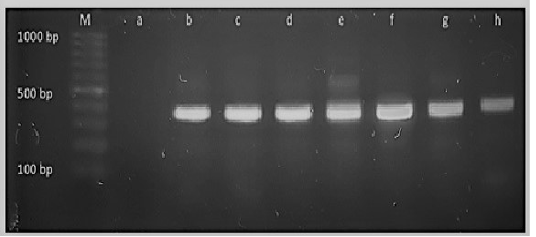
Figure 4: M: marker, Lane a: Negative control, Lane b: Positive control Lanes c-h: Positive amplification of 16Sr DNA gene at 375bp
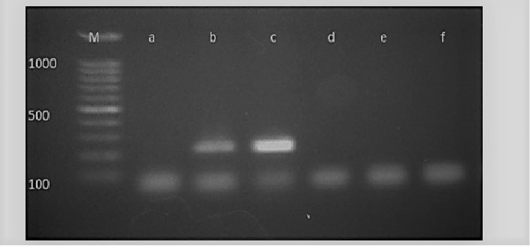
Figure 5: M: marker, Lane a: Negative control, Lane b: Positive control, Lanes c: Positive amplification of nucA gene at 239 bp and Lane d-f: Negative amplification of nucA gene
After phenotypic examination; biochemical tests and molecular identification of bacterial isolates, results in Table (5) revealed that S.aureus was isolated from 26.92% of all
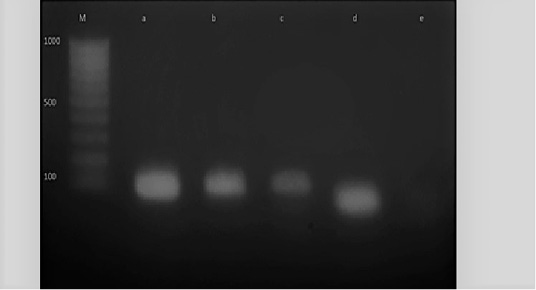
Figure 6: M: marker, Lane e: Negative control, Lane a: Positive control, Lane b and c: Positive amplification of nuc gene at 126 bp and Lane d: Negative amplification of nuc gene
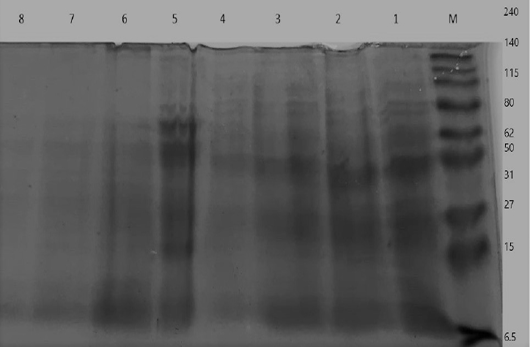
Figure 7: Electrophoretic whole cell protein profiles of different isolated bacteria Lane 1: Molecular size marker (M) with mol. wt. ranges from (6.5 to 240) KD. ; Lanes (1- 4): Staphylococcus bacterial isolates, Lanes (5-8): isolated SCVs.
examined milk samples and from 2.38% of normal milk samples, 35.53% of subclinical mastitic milk samples and 58.33 % of clinical mastitic milk samples. Prevalence of SCVs S.aureus was 13.85% and they were only isolated from subclinical mastitic milk sample which mostly were chronic cases with percentage of 23.68 % but not isolated from either normal or clinical milk samples. Other Staphylococcus spp. were isolated from 16.15% of all examined milk samples and from 9.52%, 19.74% and 16.67% of normal, subclinical and clinical milk mastitic samples, respectively.
UPGMA clustering dendrogram analysis was used to compare the protein fingerprints of the isolated SCVs S.aureus with protein fingerprints of parent S.aureus isolates. The present dendrogram proved that there were high to moderate similarities in the protein fingerprints of the different parent S. aureus isolates (from 0.40 to 0.71), i.e. approximately (from 40% to 71%) when converted to percent values. At the same time, the protein fingerprints of the different SCVs S.aureus isolates showed somewhat high to moderate similarities (from 0.38 to 0.63), i.e. approximately from 38% to 63%. On the other hand, the percentages of similarities between the protein fingerprints of the isolated SCVs S.aureus and protein fingerprints of the other parent S.aureus isolates showed no or low levels of similarities (from 0 to 0.27), i.e. approximately from 0% to 27% as shown in Table (7) and (Figure 8).
DISCUSSION
Mastitis is the most widespread and imposes considerable economic losses in dairy cattle worldwide due to loss of milk production, cost of treatment, culling of milk due to antibiotic residues as well as cows with chronic or recurrent mastitis (Hussein et al., 2018). It is well known that, mastitis is divided into clinical with signs of inflammation in milk (blood, pus and flaks) and udder abnormalities or subclinical without visible signs (Ruegg, 2017).
In the current study the prevalence of sub-clinical and clinical mastitis was estimated depending on clinical examination of the udder and CMT on apparently normal milk samples. The prevalence of subclinical mastitis in the collected milk samples was 58.46%, most of these cases were suffered from chronic or recurrent mastitis. These results came in accordance with Abebe et al. (2016), Amer et al. (2018) and Pumipuntu et al. (2019) who recorded subclinical mastitis prevalence in 59.2%, 60% and 59%, respectively. They reported that the prevalence of subclinical mastitis was attributed to shortage of the routine mastitis prevention and control practices. On the other hand other results were less than ours (Hussein, 2012) (38.89%) and (Katsande et al., 2013) (21.1%).
Our results revealed that the incidence of clinical mastitis was 9.23%., nearly similar results were obtained by Zeryehun & Abera, (2017) and Amer et al. (2018) who found the incidence of clinical mastitis (12.5%) and (11.6 %.), respectively in their studies. While Abebe et al. (2016) and Gao et al. (2017) reported that 3.4% and 3.3% of cows had clinical mastitis, respectively which were lower than our results.
In the last decade one of most important contagious microorganism causing mastitis is Staphylococcus aureus (Graber & Bodmer, 2019). Genetic plasticity of S.aureus has enabled the rise of persistent and multidrug-resistant strains those mutants called small colony variants (SCVs) of S.aureus (Proctor et al., 2006). SCVs S.aureus have been recognized for many decades (Jensen, 1957), but only flashed more research interest in latest years when they were found to be associated with chronic recurrent infections in both humans (Masoud-Landgraf et al., 2016) as well as animals (Atalla et al., 2008) and (Alkasir et al., 2012) allowing their existence in undesirable situations (Bui et al., 2015).
Kahl et al. (2003) explained that colonies of S.aureus on mannitol salt agar that changed from orange to yellow were considered as parent or wild S.aureus. The non-hemolytic, non-pigmented, pin-pointed colonies on blood agar and small colonies on Mannitol salt agar that did not change color were considered SCVs S.aureus. Gram staining, catalase and coagulase tests were done for confirmation (Brakstad et al., 1992). At the same time, Zhu et al. (2016) recorded that colonies of SCVs S.aureus were pin-pointed (0.3mm), non-pigmented and failed to make hemolysis on blood agar until incubated at 37 OC for 24 h, and under the light microscope, these prospective SCVs appeared to be indistinguishable to typical S.aureus. While coagulase was not identified, catalase test was positive.
In our study differences in phenotypic characters between both parent S.aureus and SCVs was observed. Although, parent strains of S.aureus grew rapidly with large yellow colonies as they were able to ferment Mannitol, coagulate rabbit plasma, catalase positive and they also had hemolytic colonies on blood agar media, SCVs slowly grew up to 48 hrs forming small colorless colonies failed to ferment Mannitol, coagulase negative, catalase positive and when SCVs strains grow on blood agar often colonies were non-hemolytic, non-pigmented pinpointed and biochemical tests were frequently non-reactive. Slow growth rate and colony size of SCVs may be attributed to its auxotrophic to haemin, menadione (von Eiff et al., 2006), or thymidine (Maduka-Ezeh et al., 2012) which lead to deficiencies in energy production and tricarboxylic acid metabolism, later they showed slower in development with smaller size (Proctor et al., 2006).
In the present study as shown in table (5), 26.92% of bacterial isolates were parent S.aureus from all examined milk samples, and from 2.38% of the normal milk samples, 35.53% from subclinical mastitic milk samples and 58.33 % of clinical milk samples. Prevalence of SCVs S.aureus was 13.85% and they were only isolated from milk samples collected from cows showed infection with subclinical mastitis which mostly were chronic cases with percentage of 23.68 % but not isolated from either normal or clinically infected milk samples. Other Staphylococcus spp.; Coagulase-negative staphylococci (CNS) were isolated from 16.15% of all the examined milk samples and from 9.52%, 19.74% and 16.67% of normal milk samples, affected with subclinical and clinical mastitis, respectively. Our results nearly came in agreement with Salam, (2015) and Ndahetuye et al. (2019) who reported that S.aureus isolates were recovered from 23.5% and 22% of examined milk samples, respectively. On the other hand, higher results were obtained by Ameen et al. (2019) and Suleiman et al. (2018) who showed that the dominant bacterial species in milk was S.aureus as they were isolated from 38% and 36.8%, respectively of the examined samples. Other studies reported by Abebe et al. (2016) and Hoque et al. (2018) revealed that the prevalence of S.aureus was 51.2% and 74.0% of the examined milk samples, respectively. Mean while, lower percent were isolated by Hosseinzadeh & Dastmalchi Saei, (2014) and El-Ashker et al. (2015) who recorded that S. aureus was present in 4.4% and 5.6%, respectively of all collected samples.
The present study showed that the prevalence of SCVs S.aureus was higher than those recorded by Zhu et al. (2016) who revealed that only one strain of S. aureus SCVs was isolated from 30 tested samples of cows suffer from chronic S.aureus mastitis; whereas Atalla et al. (2008) suggested that, SCVs may be an important contributor to the prolonged survival of S.aureus in some cases of mastitis as they isolated one strain of SCVs from 6 S.aureus isolates.
Due to the difficulty and prolonged time of isolation and identification of SCVs grown by conventional methods so identification was mostly performed by 16S rRNA gene sequencing for confirmation of S.aureus SCVs (Kahl et al., 2016). Hoque et al. (2018) stated that detection of S. aureus by PCR was done through using of the thermo nuclease gene (nuc; S.aureus specific gene). In this study three primers were used (16S rDNA, nuc and nucA) for genotypic identification of SCVs after phenotypic characterization as described by Zhu et al. (2016).
The current results revealed that, the prevalence of parent S.aureus and SCVs S.aureus was 26.92% and 15.38%, respectively (Table 4) by using phenotypic characters and biochemical tests. While the prevalence of SCVs S.aureus was 13.84% by PCR that means 2 isolates were misdiagnosed by phenotypic detection. Closely similar results were stated by Zhu et al. (2016) who recorded that the potential SCVs were identified as S.aureus SCVs with the genes (nuc, nucA, and 16S rDNA) by multiple PCR amplification and all isolates were positive to multiple PCR test. They added that by the sequence analysis of 16S rRNA, the gene of S.aureus SCVs and its parental strains were homologous with published S.aureus sequence. Consequently, Brakstad et al. (1992) also stated that, the detection of nucA by PCR was useful for identification of SCVs S.aureus. Furthermore, He et al. (2009) combined between the observation of colony phenotype and identification of the species-specific gene nuc of S. aureus by PCR amplification to confirm isolation of SCVs.
Kapoor et al. (2017) stated that antimicrobial therapy is an important strategy for mastitis control. However, imprudent use of antibiotics, may lead to a rise in antimicrobial resistance and there is hazard of later spread of resistant genes to other microbial populations. High incidence of antibiotic resistance was commonly detected in both bovine and human S.aureus isolates from Egypt (El-Jakee et al., 2011). The ongoing work revealed that SCVs S.aureus were more resistant to antibiotics than parent S.aureus isolates, as illustrated in Table (6). The isolated SCVs S.aureus strains were susceptible to Ciprofloxacin (72.22%), Enrofloxacin (66.67%) and Rifampin (88.90%) while they had different resistances to almost all other used antibiotics. On the other side, S.aureus isolates were sensitive to Ciprofloxacin (80%), Enrofloxacin (85.71%), Amoxicillin-clavulanic acid (68.57%), Florofenicol (71.42%), Gentamycin (62.86%), Rifampin (80%), Cefoquinom (88.57%) and trimethoprim-sulfamethoxazole (54.29%) and they had high resistance to Amoxicillin (91.43%), Penicillin (74.29%), Tetracycline (80%), Streptomycin (77.17%) and Kanamycin (65.71%).
Our results clarified that SCVs S.aureus were more resistant to antibiotics than parent S.aureus, and these findings coincided with several other studies as Sendi & Proctor, (2009), which showed that SCVs show increased resistance to intracellular defenses and reduced stimulation of host defenses compared to ‘normal’ staphylococcal strains, and this can cause latent or recurrent infections, Jonsson et al. (2003) exposed that SCVs were more virulent than its isogenic parental strain in the model of septic arthritis and have been shown to be more resistant to antibiotics due to the ability of SCVs to produce high amounts of destructive proteases.
However, Ou et al. (2016) indicated that SCVs have been shown to shut off their development of toxin within the cell allowing them to live for long periods of time in the host without inducing an immune response. The transition from wild type S.aureus to SCVs was therefore strongly correlated with improved survival within the host and protection against antibiotics and host immune cells (Tuchscherr et al., 2010).
On the other hand, von Eiff et al. (2006) illustrated that SCVs increased resistance to amino glycosides may attributed to the low content of ATP in SCVs which causes incompetent transport of aminoglycosides into the cell, moreover the intracellular situation might safeguard SCVs from host defenses and antimicrobial agents, thereby providing one explanation for the struggle in clearance SCVs from host tissue. Looney, (2000) and Sinha & Herrmann, (2005) added that likewise SCVs were more resistant to cell-wall-active antibiotics due to slow growth rate which reduce cell-wall division leading to reduction of the efficacy of β-lactam antibiotics, and they also can survive within professional and nonprofessional phagocytic cells and subsequently restrain immune responses.
Atalla et al. (2008) recorded that, SCVs were resistant to gentamicin but was almost as sensitive as other strains to rifampicin, which is simulated our results. Recently, Zhu et al. (2016) recorded nearly similar results as they recorded that, SCVs S.aureus had a high resistance to SXT, while their parental strains were sensitive, S.aureus isolates were sensitive to Oxacillin, Ampicillin, Streptomycin, Gentamicin and SXT, while SCVs were resistant to SXT, Streptomycin, Ampicillin and Gentamicin which go along with our findings.
Some studies showed that for optimum eradication of SCVs, treatment should be made of therapy with an antibiotic that has best bactericidal activity against slow-growing bacteria, although Rifampin has very good bactericidal activity against slow-growing bacteria, it should be given in combination with other antibiotics due to the rapid development of resistance when given unaccompanied (Vaudaux et al., 2006). The combination suggested by Sendi et al. (2006) rifampin with a fluoroquinolone, has been shown to be effective for treatment of hip replacements in persons infected with S.aureus SCVs.
Examination of whole-cell protein profiles by SDS–PAGE has lately been well-known as a valuable method for the identification of a number of bacteria (Elliott & Facklam, 1993). In the same time provided valuable epidemiological information that could be used in isolation of the microorganism (Berber et al., 2003).
The one-dimensional SDS-PAGE and the UPGMA (Table 7) & (Figure 7 & 8) clustering dendrogram analyses were done to compare the protein fingerprints of the isolated SCVs and that of parent S.aureus bacterial isolates. Dendrogram analysis illustrated that there were somewhat high to moderate similarities in the protein fingerprints of the different S.aureus isolates from 40% to 71%. At the same time, the protein fingerprints of the different SCVs S.aureus isolates showed somewhat moderate similarities from 38% to 63%. On the other hand, the percentages of similarities between the protein fingerprints of the isolated SCVs and protein fingerprints of parent S.aureus isolates showed no or low levels of similarities from 0% to 27% as shown in Table (7) and (Figure 8).
The absence or weak similarities between the isolated parent S.aureus strains and SCVs S.aureus may be attributed to the great variation in gene content which may be caused by the recurrent treatment with antibiotics or it may be referred to different environmental stressors or probably have contributed to the degree of virulence even within similar clonal genotype (Atshan et al., 2015). Likewise, results of Berber et al. (2003) showed that whole-cell and extracellular protein profiles differed in several protein bands in Staphylococcus aureus and other species of Staphylococcus, and that, differences were detected between protein profiles of S. aureus strains and other Staphylococci strains. Finally, they accepted that the comparison of electrophoretic protein patterns offers a dependable measure of genomic relatedness. Given this setting, it is very hopeful that, upon quantitative analysis of whole-cell and extracellular protein profiles, the strains of Staphylococcus spp. formed clusters with at least 90% similarity.
Our findings agreed with the results of Costas et al. (1989) who stated that, they were able to group the MRSA (methicillin-resistant Staphylococcus aureus) strains objectively and reproducibly into four phenons when they investigated their typing using SDS-PAGE of proteins. They added, the underlying pattern of bands may represent «fingerprints,» and it may be possible to define additional groups and thus increase the discriminatory power of the method.
CONCLUSION
Results showed the importance of SCVs S.aureus in dairy herd health as it causes chronic or recurrent subclinical mastitis which lead to more economical losses due to misdiagnosis as well as lack of treatment. Due to difficulty of diagnosis the percentage of SCVs S.aureus may increase so these subpopulations must be actively sought in the routine diagnosis and more attention should be paid for the investigation of all types of observed colony morphologies, and isolates should be subjected to antimicrobial susceptibility testing with a protocol for treatment as it has resistance against many antibiotics. This study suggests that Ciprofloxacin, Enrofloxacin and rifampin can be used for treatment of SCVs S.aureus.
acknowledgements
The work was inancially supported by both the Animal Reproduction Research Institute (ARRI), Agricultural Research Center and National Research Centre (NRC), as a part of the project No.11020303 - the 11th research plan.
Conflict of interest
The authors declare that there is no conflict of interest.
authors contribution
All authors shared the ideas and writing of the manuscript. SSE diagnosed cases, collected milk samples and did bacteriology and sensitivity tests, MKE worked on DNA extraction and molecular diagnostic tests (PCR) and IMG worked on Protein Profile using Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis.
REFERENCES