Whole-Genome Sequencing Mapping of Candidate Genes of Cervus Nippon Hortulorum, Presumed to be Sika Deer from the Korean Peninsula
Whole-Genome Sequencing Mapping of Candidate Genes of Cervus Nippon Hortulorum, Presumed to be Sika Deer from the Korean Peninsula
Yong Su Park1a, Dong Won Seo2a, You Sam Kim2, Myung Hum Park2, Min Jee Oh3, Sang Hwan Kim3,4*
The Cervus nippon, which is thought to have been distributed in Korea, China, and Russia, was estimated to be a subspecies of C. n. hortulorum (McCullough 2009; McCullough et al., 2009; Saggiomo et al., 2020).
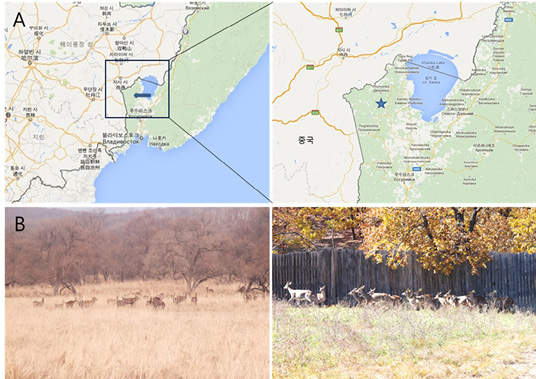
Photographs of the Sika deer sampling site and Sika deer farm used in this study; A: Sika deer sampling site; B: Farm where Sika deer are raised.
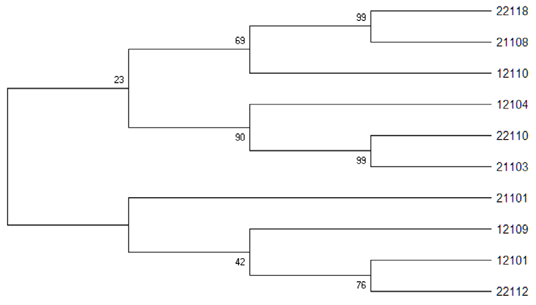
Phylogenetic tree result using CST2 primer set for 10 Korean native deer.
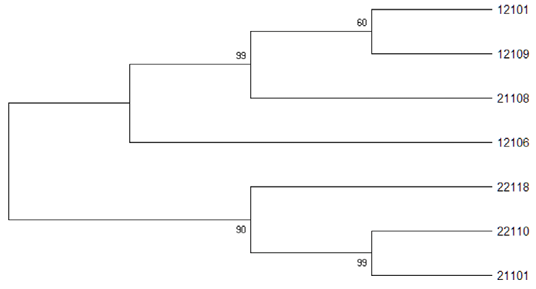
Phylogenetic tree result using CST39 primer set for 7 Korean native deer.