Single Nucleotide Polymorphism Induces a Positive Selection Pressure at Gag-Pol Sites in Human Immunodeficiency Virus Favoring Drug Resistance Mutations
Single Nucleotide Polymorphism Induces a Positive Selection Pressure at Gag-Pol Sites in Human Immunodeficiency Virus Favoring Drug Resistance Mutations
Amreen Zahra1, Mushtaq A. Saleem1*, Hasnain Javed2, Muhammad Azmat Ullah Khan3, Muhammad Naveed1 and Abdul Rauf Shakoori4
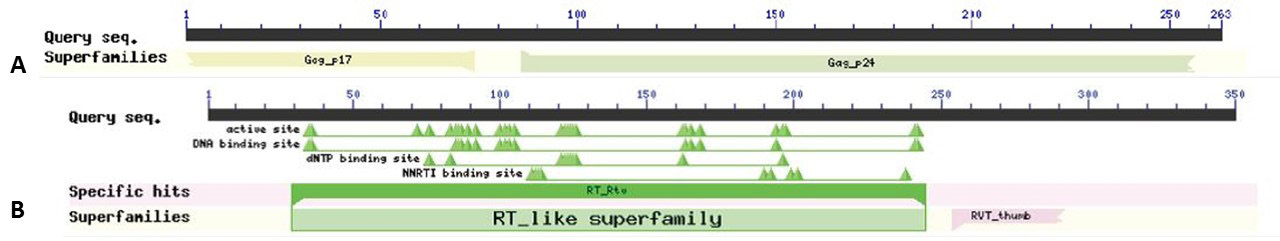
Sequence matches and features of Gag-Pol-pol as predicted by Pfam: 10 domains Gag_p17, Gag_p24, zf-CCHC, RVT-1, RVT_connect, RNase-H, RVP, RVT-thumb, MLVINC, Integrase-Zn were best predicted by Pfam as shown in the figure.
A and B, Predicted conserved domains and E-values of Gag-Pol by CDD database of NCBI: Highly conserved regions of Gag_p17, Gag_p24, RVT-1, RVT _thumb were predicted in HIV-1.The domains for Pol showed an active site, DNA binding site, dNTP binding site and NNRTI binding site.
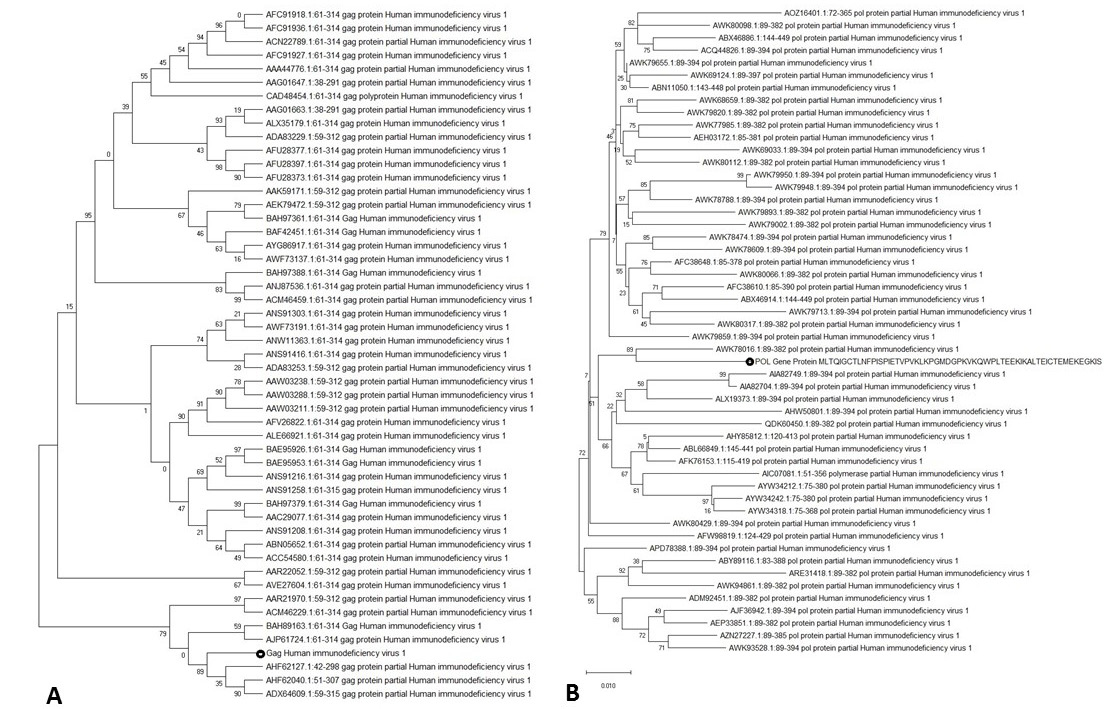
Phylogenetic tree generated via Neighbor Joining method. A: shows phylogenetic tree of Gag. B: shows phylogenetic data of Pol.
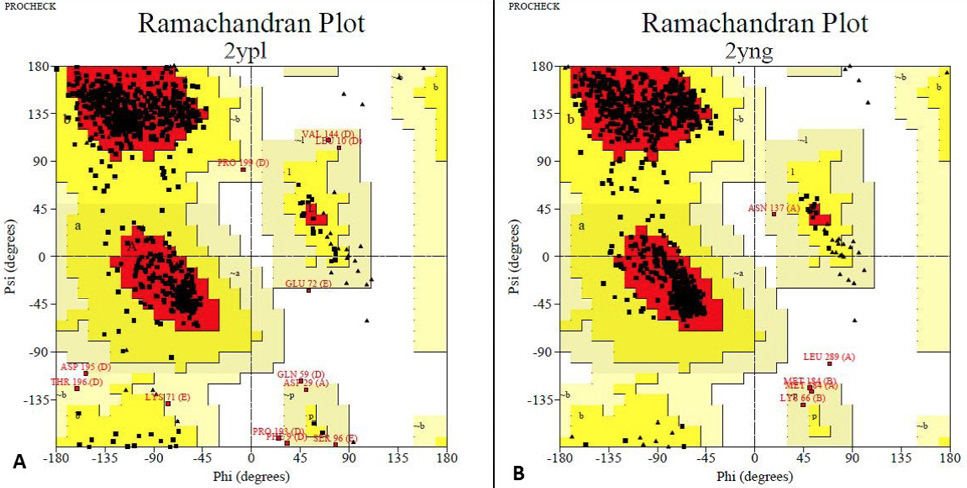
Rampage analysis: The Ramachandran plots show the phi (ϕ)-psi (ψ) torsion angles for every residue of a protein. A, The final model had 87.3% residues in the most favored regions, 11.3% in additional allowed regions. Total number of residues were 821. B, The final model had 92.5% residues in the most favored regions, 6.9 % in additional allowed regions. Total number of residues were 971. Based on an analysis of 118 structures of resolution of at least 2.0 Angstroms and R-factor no greater than 20%, a good quality model would be expected to have over 90% in the most favoured regions.
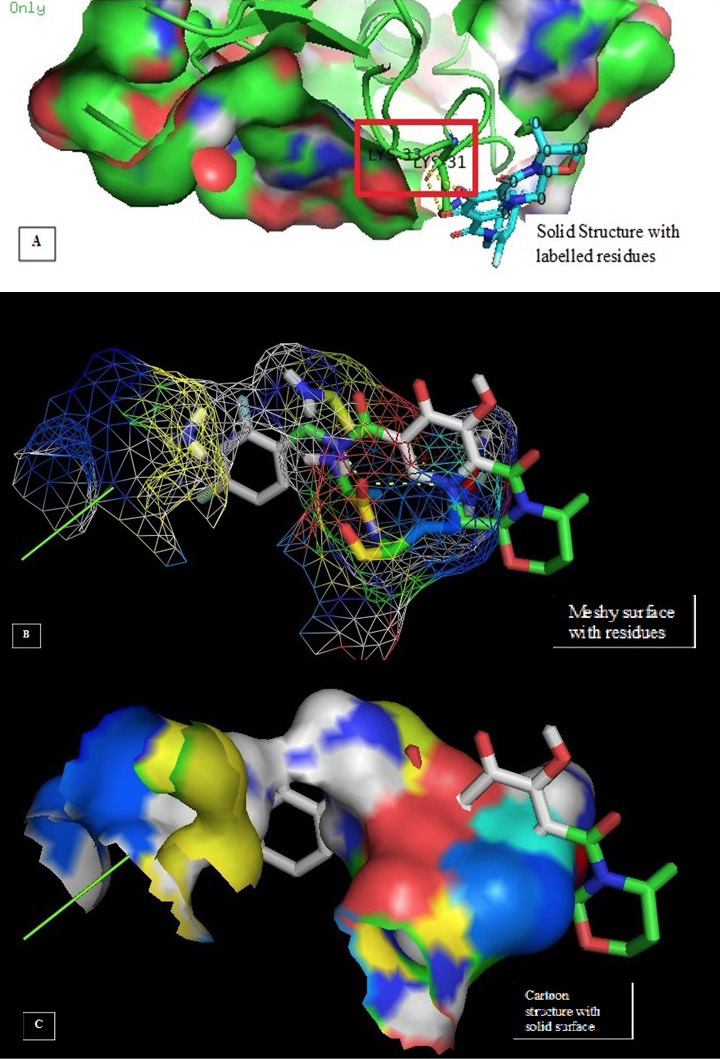
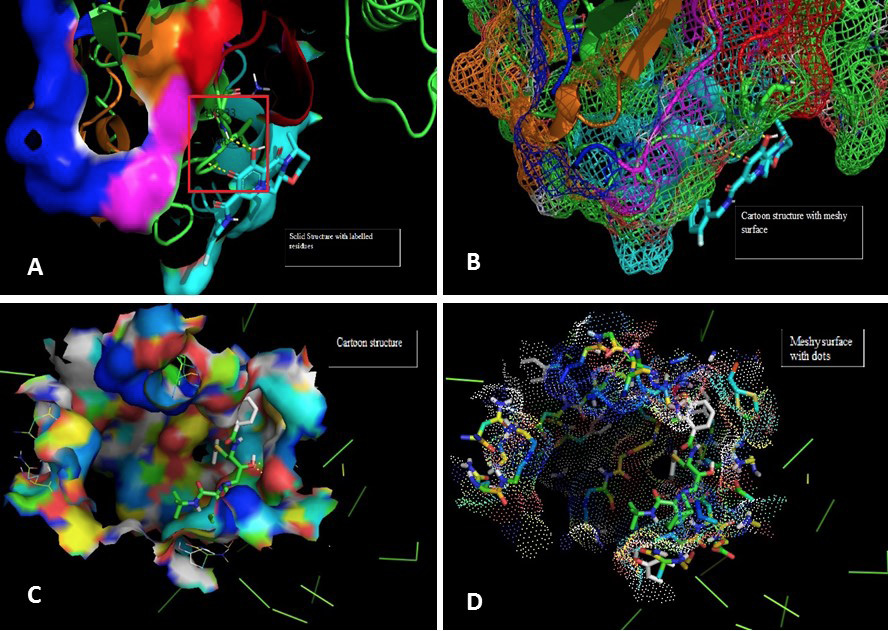
Molecular docking image analysis of Gag. The interactions are visualized between the protein and ligand of Dolutegravir for Gag protein. Dolutegravir is one of the core drugs used for treatment of HIV-1 targeting the Gag-Pol region. A shows the solid structure with residues; B represents the meshy surface and C represents the solid surface cartoon.
Molecular docking image analysis of Pol. The interactions are visualized between the protein and ligand of Dolutegravir with Pol protein. A shows the solid surface of the protein with residue labels Lysine and valine of amino acids. B represents the cartoon structure with meshy surface depicting the binding interaction of different residues of receptor with ligand atoms as shown. In figure C the cartoon structure and binding yellow lines show the hydrogen bonds. D demonstrates the meshy surface with dots.