Reliable Scarless Fusion of Genes through Overlap Extension PCR to Develop Chimeric Genes
Reliable Scarless Fusion of Genes through Overlap Extension PCR to Develop Chimeric Genes
Iram Gull*, Muhammad Shahbaz Aslam, Imran Tipu, Roohi Mushtaq and Muhammad Amin Athar
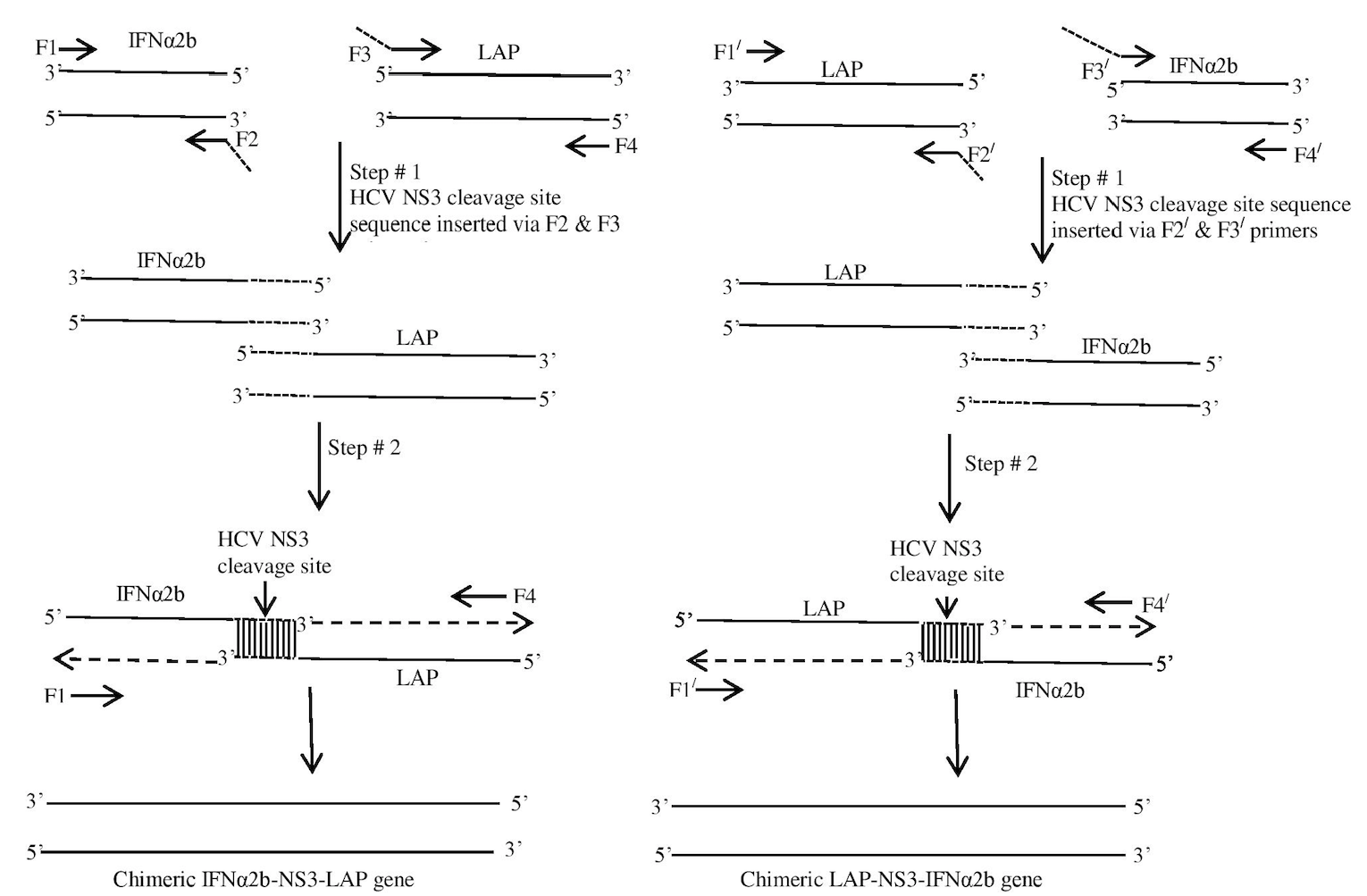
Two step overlap extension PCR for construction of chimeric gene products (IFNα2b-NS3-LAP and LAP-NS3-IFNα2b). F2, F3 and F2/, F3/ were chimeric primers that introduced the overhangs of HCV NS3 protease cleavage site sequence when used with non-chimeric primers F1, F1/ and F4, F4/ in step # 1 of SOE-PCR. Overhangs introduced overlapping regions that act as primer in initial 10 cycles of second step of PCR. First 10 cycles of step # 2 were performed without addition of any primer and strands were extended from 3’ ends of the overlapping region. In the later 30 cycles, non-chimeric primers F1/F4 and F1//F4/ were added in reaction vial to amplify the full length chimeric gene products.
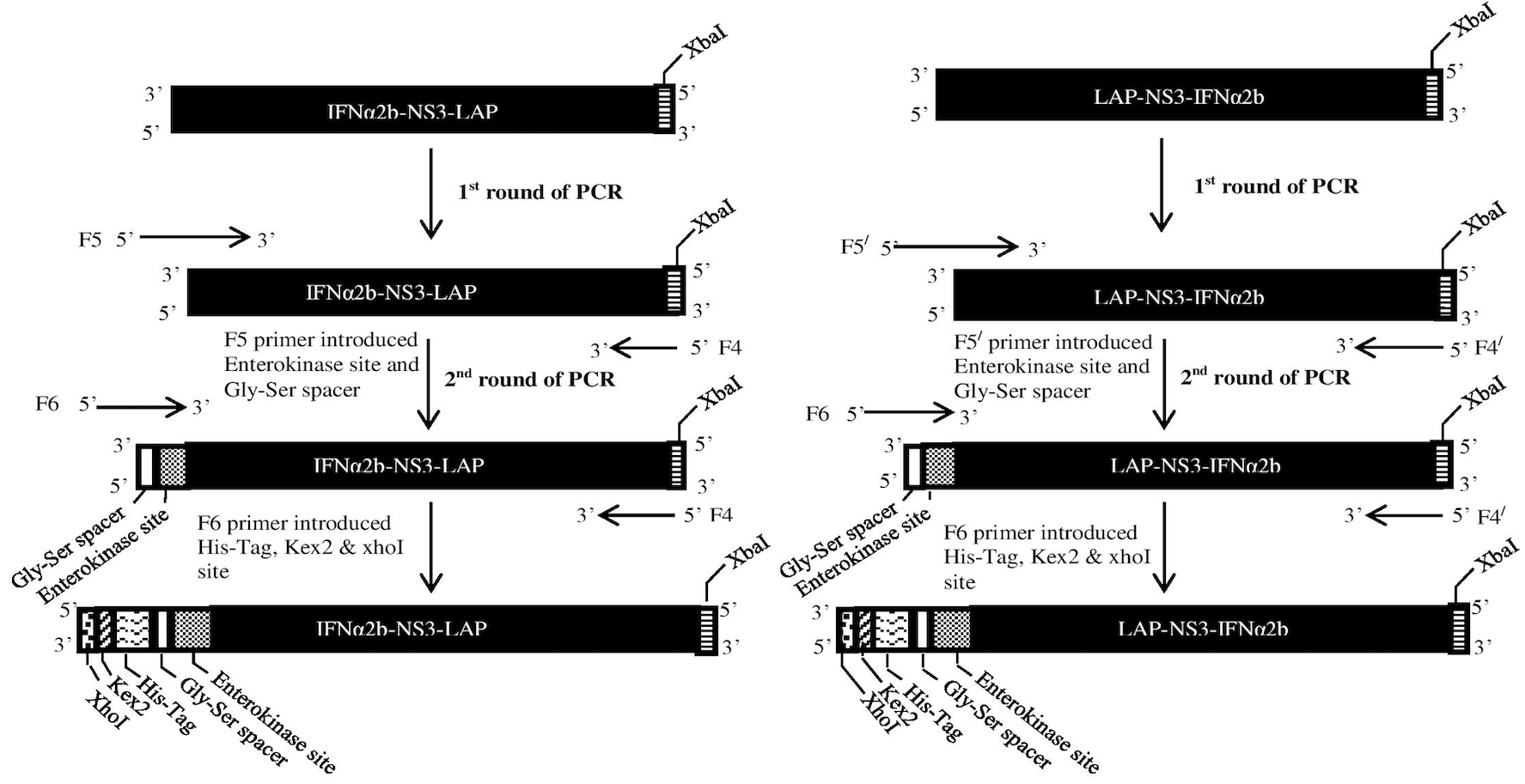
Overlap Primer Walk PCR (OPW-PCR). In 1st round of PCR, Primer F5 and F5/ were used to introduce enterokinase cleavage site at 5’ end of chimeric gene products IFNα2b-NS3-LAP and LAP-NS3-IFNα2b respectively. In 2nd round of PCR, primer F6 introduced Gly-Ser spacer, His-Tag, Kex2 and XhoI restriction site at 5’ end of the PCR product of 1st round.
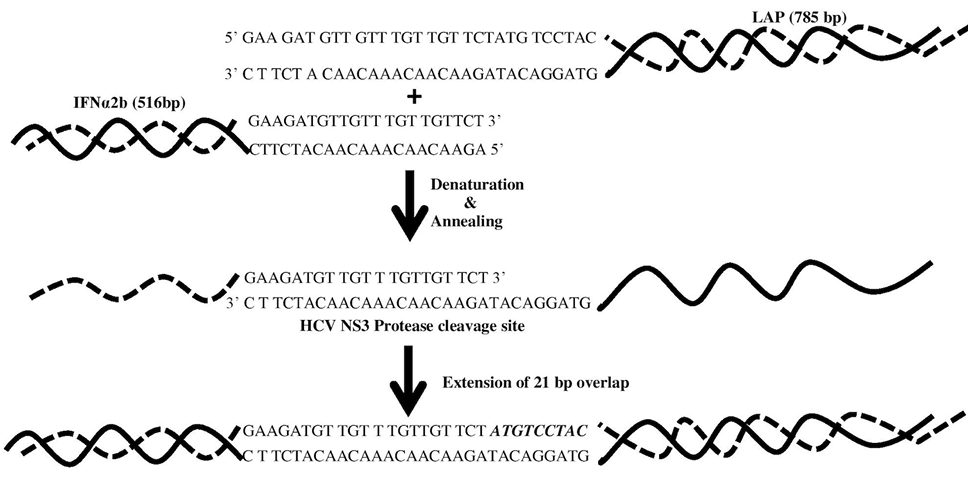
Splicing during the first 10 cycles of step # 2 of SOE- PCR driven without addition of primers. Inserted overlapping region of HCV NS3 protease cleavage site sequence acts as primers for strands extension. HCV NS3 protease cleavage site sequence is highlighted by alphabets.
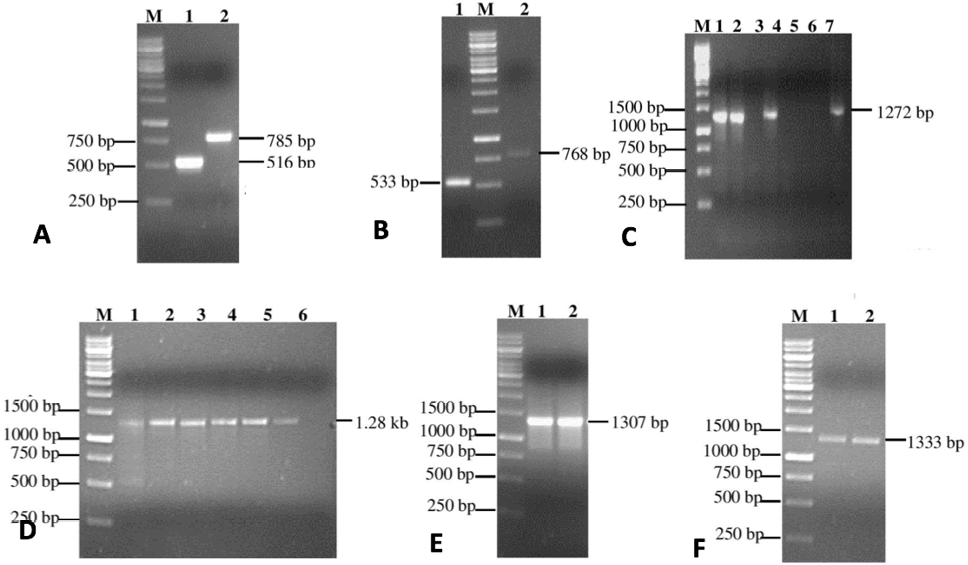
Analysis of PCR products by 1% agarose gel electrophoresis. Gels were visualized by staining with ethidium bromide; A, Gel purified PCR products of steps # 1 of SOE-PCR. Lane M: DNA size marker, Lane 1: IFNα2b gene (516 bp) amplified using primer pair F1 and F221, Lane 2: LAP gene (785 bp) amplified using primer pair F3 and F4; B, Gel purified PCR products of steps # 1 of SOE-PCR . Lane M: DNA size marker, Lane 1: IFNα2b gene (533 bp) amplified using primer pair F3/ and F4/, Lane 2: LAP gene (768 bp) amplified using primer pair F1/ and F2/21; C, Optimization of annealing temperature (44-60 ͦ C) in first 10 cycles of step # 2 of SOE-PCR during which 30 bp overhang (HCV NS3 protease cleavage site sequence ) at the 3’ end of IFNα2b gene (516 bp) overlap by 21 bp with 5’ end overhang of LAP gene (774 bp). In later 30 cycles, 58 °C annealing temperature was used with primer pair F1 and F4 to get 1.28 kb chimeric gene product. Lane M: DNA size marker, Annealing temperatures in Lane 1: 50 ͦ C, Lane 2: 55 ͦ C, Lane 3: 60 ͦC, Lane 4: 58 ͦ C, Lane 5: 48 ͦ C, Lane 6: 44 ͦ C and in Lane 7: 58 ͦ C; D, Optimization of minimum required equimolar concentration of genes for SOE-PCR. Lane M: DNA size marker, PCR product using equimolar concentration in Lane 1: 0.80 pmoles, Lane 2: 0.20 pmoles, Lane 3: 0.10 pmoles, Lane 4: 0.05 pmoles, Lane 5: 0.025 pmoles, Lane 6: 0.0125 pmoles and Lane 7: 0.00625 pmoles; E, 1st step of OPW-PCR with F5/F4 and F5//F4/ primer using chimeric genes IFNα2b-NS3-LAP and LAP-NS3-IFNα2b respectively introduced enterikinase cleavage site and Gly-Ser spacer at 5’ end; F, 2nd round of OPW-PCR with F6/F4 and F6/F4/ primer using PCR products of 1st round of OPW-PCR as templates to introduce His-Tag sequence and XhoI cleavage site.