Polymorphism of Kappa-Casein Gene (CSN3|Alw44I) and Its Effect on Milk Yield and Compositions in Indigenous Senduro Goat
Research Article
Polymorphism of Kappa-Casein Gene (CSN3|Alw44I) and Its Effect on Milk Yield and Compositions in Indigenous Senduro Goat
Tri Eko Susilorini1*, Ahmad Furqon2, Wike Andre Septian1, Desinta Wulandari3, Suyadi Suyadi3
1Department of Animal Production, Faculty of Animal Science, Universitas Brawijaya Malang, East Java 65145, Indonesia; 2National Research and Innovation Agency, Republic of Indonesia, Bogor, West Java, Indonesia; 3Department of Animal Reproduction, Faculty of Animal Science, Universitas Brawijaya Malang, East Java 65145, Indonesia.
Abstract | Casein is one of the major milk proteins approximately 80-83% of total protein. The CSN3 gene has been broadly studied due to its important influence on the milk properties. Senduro goat is local breeds in Indonesia that provide milk. This study was aimed to analyse CSN3 gene polymorphism and its association with milk production and composition on Senduro goat. A total of 42 lactating Senduro goat were used in study from parity 1 to 4. Blood and milk samples were collected in Senduro district, Lumajang. The traits observed were milk yield, fat, lactose, solid non-fat, total-solid, and protein. The CSN3|Alw44I polymorphism was analysed using Polymerase Chain Reaction-Restricted Fragment Length Polymorphism method. The result showed that three genotypes (CC, CT, and TT) and two alleles (C and T) were found. The frequencies of CC, CT, and TT genotypes were 0.450, 0.500, and 0.050 respectively, while the frequencies of C and T allele were 0.702 and 0.298, respectively. The different genotypes didn’t significantly affect milk production and compositions. Descriptively, Senduro goat with TT genotype had the highest milk production and compositions. In conclusion, the polymorphism of CSN3|Alw44I was polymorphic and could differentiate milk production and compositions among genotypes (TT>CT>CC).
Keywords | Key words: PCR-RFLP, Restriction enzyme, Polymorphic, Local goat, Kappa-casein
Received | March 24, 2022; Accepted | April 23, 2022; Published | June 15, 2022
*Correspondence | Tri Eko Susilorini, Department of Animal Production, Faculty of Animal Science, Universitas Brawijaya Malang, East Java 65145, Indonesia; Email: triekos@ub.ac.id
Citation | Susilorini TE, Furqon A, Septian WA, Wulandari D, Suyadi S (2022). Polymorphism of kappa-casein gene (CSN3|Alw44I) and its effect on milk yield and compositions in indigenous senduro goat. Adv. Anim. Vet. Sci. 10(6):1333-1338.
DOI | https://dx.doi.org/10.17582/journal.aavs/2022/10.6.1333.1338
ISSN (Online) | 2307-8316
Copyright: 2022 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
INTRODUCTION
Senduro goat is one of local breeds in Indonesia and commonly raised by the farmers as the dual-purpose goat. This goat has an important role to provide animal products such as meat and milk. The goat’s milk could be processed into pasteurized milk, ultra-high temperature milk, powder milk, yoghurt, cheese, butter and ice cream as dairy products (Aryana and Olson, 2017; Fazilah et al., 2018; Kalyankar and Patil, 2016). These products are important parts to fulfil the daily human diet. Recently, the goat’s milk is well known as the functional food due to nutritional properties and health promoting compounds, such as oligosaccharides, conjugated linoleic acids, and bioactive peptides or protein (Salva et al., 2011; Abeijón Mukdsi et al., 2013).
Casein is one of the major protein of milk approximately 80-83% of total protein and consist of alpha (α), beta (β), and kappa (ĸ) casein (Fan et al., 2020). Gipson (2019) reported that casein is the main protein in goat milk. Casein provides the body with all of the amino acids necessary to help build muscle and the main constituent of milk protein. The casein protein coding region have been mapped on chromosome 6 with coding genes CSN1S1 (αs1-casein), CSN1S2 (αs2-casein), CSN2 (β-casein), and CSN3 (ĸ-casein) in goat (Rehman et al., 2021). CSN3 have an important role to influence the size of milk micelles, which has an impact on the coagulation features of the milk in cheese making process (Ahmed, 2011). The CSN3 (ĸ-casein) gene has been broadly studied due to its influence on the properties of milk. Kappa casein is most important protein and it is being determined at chromosome number BTA6 that consists of five exons spread over about 13.06 kb and exon 4 is most of the protein-coding region (Akyuz et al., 2012; Volkandari et al., 2017; Barbosa et al., 2019). CSN3 gene in goat is highly polymorphic in several studies (Gupta et al., 2009; Kiplagat et al., 2010). Kumar et al. (2009) reported that there are 15 polymorphic sites in the exon 4 of CSN3 gene. Some variants were reported to have a significant effect on the production and nutritional composition of the milk (Caravaca et al., 2009, 2010).
The milk yield and composition in goats were reported have high variability (Gautam et al., 2020), that might be influenced by genetic variability (Singh et al., 2018). The selection for major genes will be profitable in efficient breeding scheme (Mucha et al., 2018). CSN3 was selected to support goat breeding program due to its effect on the milk and highly polymorphic gene. In addition, there needs to be an association between genotype and milk production. This study was aimed to analyse Kappa Casein gene (CSN3) polymorphism and its association with milk production and compositions on Senduro goat.
MATERIALS AND METHODS
Animal and phenotypic parameters
Senduro goat is a dairy goat type that is mostly kept on the slopes of the Semeru Mountains, which includes 8 villages in Senduro District, Lumajang Regency, East Java. This area has a cold-humid climate with an annual temperature range of 25–27oC and humidity of 80-90%. The total population of Senduro goats almost reached 10,000 in 2018. This goat has an important role in supporting local economic society (Arsa, 2018).
All procedures related to animal use in this study were approved by the Animal Care and Use Committee of Universitas Brawijaya under regulation number 020-KEP-UB-2021 (Ethical Clearance). A total of 42 lactating Senduro goat aged 2-4 years were used in this study. The goats had parities ranged from 1 to 4. All goats were reared by farmers intensively. Milk yield was visually measured using the measuring cup of 1000 and 500 mL. Milk compositions (fat, lactose, solid non-fat, total solid, and protein) were analyzed using Lactoscan Milk Analyzer (MCC- Milk Analyzer) in Laboratory of Dairy Science, Faculty of Animal Science, Universitas Brawijaya. Genomic analysis was performed in Laboratory of Animal Biotechnology, Faculty of Animal Science, Universitas Brawijaya.
Blood samples and DNA Isolation
Blood samples were collected from jugular vein using blood vacutainer containing EDTA. The DNA was isolated using Genomic DNA Mini Kit for blood and cultured cell (Geneaid Biotech Ltd., China). The procedures of DNA isolation followed the manufacture instructions. The quality of isolated DNA was checked using 1.5% agarose gel electrophoresis while the quantity was measured using NanoDrop ND-1000 Spectrophotometer (Thermo ScientificTM, Massachusetts, USA) respectively. The DNA concentration was adjusted to 50-100 ng/µL and stored at -20oC for the further analysis.
Gene polymorphism
The CSN3 gene polymorphism was analyzed using PCR-RFLP method in this study. A pair of primers were used in this study consisted of forward primer (F: 5’- GTA TGT GCT GAG TAG GTA TC -‘3) and reverse primer (R: 5’- CCT CTT TGA TGT CTC CTT AG -‘3). These primers were designed to amplify a 455 bp fragment of CSN3 gene according to the goat genomic sequence in GenBank (accession number NM_001285587). The DNA amplification used the mixture of 1 µL DNA template (50-100 ng/µL) and 30 µL PCR premix. The PCR premix consisted of 0.3 µL primer (10 pmol/ µL), 15 µL Go Taq Green Master Mix (Promega, USA), and 14.4 µL Nucleus Free Water (NFW). The DNA amplification was performed in Bio-Rad T100™ Thermal Cycler (Bio-Rad, USA) and the machine conditions were shown in Table 1. The PCR products were checked using electrophoresis for 35 min at 100V on 1.5% agarose gel. The PCR products were visualized under blue light of Glite 965 GW imaging system (Pacific Image Electronics Co., Ltd.) after the gel was stained using Diamond™ Nucleic Acid Dye (Promega, USA).
Table 1: The condition of thermal cycler machine during PCR.
|
PCR Step |
Temperature (oC) |
Time |
Number of cycle |
|
Pre-denaturation |
95 |
5 minutes |
Once |
|
Denaturation |
95 |
30 s |
35 times |
|
Annealing |
60 |
45 s |
|
|
Extension |
72 |
1 minute |
|
|
Final extension |
72 |
5 minutes |
Once |
Furthermore, the PCR products were digested by a restriction enzyme of Alw44I. The Alw44I restriction enzyme has a specific restriction site on G|TGCAC. A total of 5 µL PCR product was mixed with 0.7 μL of 10 × reaction buffer, 4 units of Alw44I (Thermo Fisher Scientific, Massachusetts, US) and 0.9 μL nucleus free water (NFW). The mixed solution was immediately incubated in 37oC for 10-14 hr. Genotyping was performed based on the band pattern of digested products on 2% electrophoresed agarose gel.
Data analysis
Genotype and allele frequency
The genotype and allele frequency were calculated using the following formula of (Nei and Kumar, 2000):
Genotype frequency:
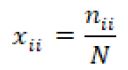
Allele frequency:
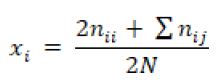
Where χii was frequency of ii genotype (CC, CT and TT); χi was frequency of i allele (C and T); nii was number of individuals with ii genotype; nij was number of individuals with ij genotype; N was number of samples.
Hardy-Weinberg equilibrium (HWE)
The Hardy-Weinberg equilibrium was calculated using Chi-square test (Hartl and Clark, 1997).
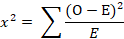
Where χ2 was HWE test; O was the observed number of genotypes; E was the expected number of genotypes; α was 5% significance level; df was number of genotype probabilities – number of alleles.
CSN3 gene association with milk yield and compositions
The association study between CSN3 gene polymorphism and milk compositions was analyzed using General Linear Model procedure in SPSS ver. 26.0. The fixed factor was the genotypes and parities while the dependent variables were milk yield, fat, lactose, solid non-fat, total solid, and protein. The mathematical model used in this research was:
Yijk = µ+Gi+Pj+ɛijk
Where Yijk was observation value (milk yield and compositions); µ was overall mean; Gi was the effect of ith genotype (CC, CT and TT); Pj was the effect of jth parity (1, 2, 3 and 4); ɛijk was random error.
RESULTS AND DISCUSSION
The production and composition of goat’s milk are influenced by many factors, one of them is parity. Effect of parity on milk yield and compositions on Senduro goats are presented in Table 2. The result showed that the parity had no effect on milk yield and compositions on indigenous Senduro goat (P>0.05), but it showed an almost steady increasing from first to third parity in all phases of lactation on milk yield. The first parity had the lowest milk yield and the third parity had the greatest milk yield (828.30 L and 1,114.53 L, respectively).
The increasing of goat age indicated the already developed alveoli that affecting the higher milk yield. On the other hand, there is a reduction in the use of energy for growth, favoring partitioning of nutrients toward milk production (Lérias et al., 2014). Milk production will continue to increase from the first parity to the fourth parity and will decrease at the fifth parity, caused by the increasing of maturity and readiness of cells and hormonal systems associated with reproductive function in the same physiological status (Vacca et al., 2018; Ishag et al., 2012; Idowu and Adewumi, 2017; Awad et al., 2022). Zamuner et al. (2020) showed that parity did not affect the percentages of milk fat or protein of commercial dairy goats in Australia. Another research reported that increasing parity increased the total fat yield (Šlyžius et al., 2017), confirmed our present result that the highest total fat was found in the fourth parity (Table 2).
The visualization of PCR product of kappa casein gene using specific primer was a 455 bp fragment. The 455 bp fragment DNA of kappa casein gene after digested by Alw44I restriction endonuclease generated two fragments of 455 and 398 bp. Two fragments of 455 and 398 bp represented CT genotype and one fragment of 455 bp represented CC genotype while one fragment of 398 bp represented TT genotype (Figure 1). Kappa casein gene of local Tunisian goats using the Alw441 enzyme was found at 459 bp and their restriction fragments were 459 and 381 bp (Jemmali et al., 2013). While on Zawaladi goats using HaeIII was at 645 bp and their restriction fragments were 416 and 229 bp (Patel et al., 2011).
Table 2: Effect of parity on milk yield and compositions on Senduro goat.
|
Traits |
Parity 1 (n=4) |
Parity 2 (n=23) |
Parity 3 (n=11) |
Parity 4 (n=4) |
Sig. |
|
Fat (%) |
6.39±1.50 |
6.88±2.24 |
6.18±1.10 |
7.33±2.56 |
Ns |
|
Lactose (%) |
4.88±0.33 |
4.80±0.37 |
4.71±0.27 |
4.49±0.24 |
Ns |
|
SNF (%) |
8.50±0.49 |
8.27±0.71 |
8.18±0.45 |
7.67±0.53 |
Ns |
|
Solid (%) |
14.88±1.90 |
15.14±1.98 |
14.36±1.38 |
15.00±2.32 |
Ns |
|
Protein (%) |
4.54±0.24 |
4.53±0.58 |
4.55±0.24 |
4.52±0.51 |
Ns |
|
Milk yield (L/day) |
828.30±196.35 |
878.34±515.69 |
1,114.53±466.66 |
1,087.73±104.24 |
Ns |
Note: n: number of samples; Ns: not significant (P>0.05).
Table 3: Frequencies of genotypes and alleles of CSN3|Alw44I in Senduro goat.
|
Breed |
N |
Genotype frequencies |
Allele frequencies |
χ2 value |
|||
|
CC |
CT |
TT |
C |
T |
|||
|
Senduro goat |
42 |
0.450 |
0.500 |
0.050 |
0.702 |
0.298 |
0.162ns |
Note: n: number of samples; χ2 (0.05;1) = 3.84; ns: not significant.
Genotype frequencies of kappa casein gene of Senduro goat in this study are 45.0%, 50.0%, and 5.0% for CC, CT and TT respectively while allele frequencies were 0.702 for C and 0.298 for T. The C allele was observed of high frequency compare to the T allele of CSN3 in this study (Table 3). The results of this study are in accordance with the results of Orobica dairy goat in alpine valley of the Northern Region Italy for the locus of CSN3 gene with the genotype frequencies of 51.2%, 37.7% and 11.1% for AA, AB and BB, respectively with allele frequencies of 0.72 for A and 0.28 for B (Chiatti et al., 2007). The results of this study are contrast to the result from Jemmali et al. (2013) in local Tunisian goats with genotypes frequency were 12.5% AA, 27% AC, and 60.5% CC. The genotype frequencies for Sarda goats were 41.5%, 39.0%, and 7.5% for AB, BB, and AA, respectively (Pazzola et al., 2014).
Table 4: Association of CSN3|Alw44I polymorphism and milk yield and composition.
|
Traits |
CC (n = 19) |
CT (n = 21) |
TT (n = 2) |
Sig.* |
|
Fat (%) |
6.75±2.43 |
6.49±1.45 |
8.22±0.37 |
Ns |
|
Lactose (%) |
4.71±0.24 |
4.77±0.42 |
5.01±0.06 |
Ns |
|
SNF (%) |
8.13±0.62 |
8.25±0.67 |
8.54±0.08 |
Ns |
|
Solid (%) |
14.88±1.95 |
14.75±1.74 |
16.76±0.46 |
Ns |
|
Protein (%) |
4.46±0.59 |
4.52±0.4 |
4.58±0.01 |
Ns |
|
Milk yield (L/day) |
858.16± 431.47 |
1,092.48± 516.39 |
1,337.86± 136.37 |
Ns |
Note: n: number of samples; Ns: not significant (P>0.05), TT>CT>CC.
The genotypes obtained with two alleles were CC, CT and TT. This genotype was then associated with milk production and composition, as shown in Table 4. The TT genotype showed higher for milk yield and compositions compared to the CC and CT genotypes (P>0.05), TT>CT>CC. The result accordance with Wardani et al. (2022) on Senduro goat using β-lactogobulin gene that TT genotype had higher milk compositions than other genotypes. The results of Chiatti et al. (2007) showed significant association between genotypes and milk yield and composition in Orobica goats maintained in Alpine valleys (BB>AB>AA), with the same pattern of this present study. From their results they suggested that BB genotype could be used to exploit superior Orobica goat for improving milk yield, protein and casein percentages in the milk when cheese product is a selection objective.
CONCLUSIONS AND RECOMMENDATIONS
The recent findings revealed that population of Senduro goat was polymorphic and in Hardy-Weinberg equilibrium based on CSN3|Alw44I locus. Milk yield and composition are higher for the goats with genotype TT than CT and CC. Goats having T allele tend to show higher milk yield and compositions. Extensive tracing of the TT genotype is needed for Senduro goats to obtain superior genetic goats in order to support breeding programs increasing milk yield and composition so that in the future it can increase farmers’ income.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the team of Dairy Science for supporting the data and the member of Genomic and Proteomic Research Group, Faculty of Animal Science, Universitas Brawijaya for data analysis. This research was supported by research funding of Faculty of Animal Science, Universitas Brawijaya through PNBP Research Grant in 2020 Contract Number 1092/UN10F05/PN/2020.
Novelty Statement
1. Senduro goat is a local goat that until now distributed only in Senduro district, East Java, Indonesia.
2. Senduro goat is a local goat that is prioritized as a dairy goat breed in Indonesia, especially in East Java.
AUTHOR’S CONTRIBUTIONS
TES and SS idea and design. AF, WAS and DW material sample collection and lab analysis. TES, SS, AF and DW write the manuscript. SS and TES revision.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES
Abeijón-Mukdsi MC, Haro C, González SN, Medina RB (2013). Functional goat milk cheese with feruloyl esterase activity. J. Funct. Foods, 5(2): 801-809. https://doi.org/10.1016/j.jff.2013.01.026
Ahmed NS (2011). Cheese yield as affected by some parameters review. Acta Sci. Polonorum Technol. Aliment., 10: 131-153.
Akyuz B, Agaoglu OK, Ertugrul O (2012). Genetic polymorphism of kappa-casein, growth hormone and prolactin genes in Turkish native cattle breeds. Int. J. Dairy Technol., 65(1): 38-44. https://doi.org/10.1111/j.1471-0307.2011.00732.x
Arsa IMS (2018). Respon masyarakat terhadap peternakan kambing senduro sebagai potensi desa wisata di desa burno kecamatan senduro kabupaten lumajang. Faculty of Animal Science, Universitas Brawijaya. Malang. Indonesia
Aryana KJ, Olson DW (2017). A 100-year review: Yogurt and other cultured dairy products. J. Dairy Sci., 100(12): 9987-10013. https://doi.org/10.3168/jds.2017-12981
Awad MAA, Almasri OA, Ibrahim MAM, Sadek RR, Abou-Bakr S (2022). Characterization of the lactation curve in Shami cows. Adv. Anim. Vet. Sci., 10(4): 786-794. https://doi.org/10.17582/journal.aavs/2022/10.4.786.794
Barbosa SBP, de Araújo ÍIM, Martins MF, da Silva EC, Jacopini LA, Batista ÂMV, da Silva MVB (2019). Genetic association of variations in the kappa-casein and β-lactoglobulin genes with milk traits in girolando cattle. Rev. Brasil. Saude Prod. Anim., 2019: 20. https://doi.org/10.1590/s1519-9940200312019
Caravaca F, Ares JL, Carrizosa J, Urrutia B, Baena F, Jordana J (2010). Effects of α s1-casein (CSN1S1) and κ-casein (CSN3) genotypes on milk coagulation properties in Murciano-Granadina goats. J. Dairy Res., 78: 32-37. https://doi.org/10.1017/S002202991000083X
Caravaca F, Carrizosa J, Urrutia B, Baena F, Jordana J, Amills M (2009). Short communication: Effect of α S1-casein (CSN1S1) and κ-casein (CSN3) genotypes on milk composition in Murciano-Granadina goats. J. Dairy Sci., 92: 2960-2964. https://doi.org/10.3168/jds.2008-1510
Chiatti F, Chessa S, Bolla P, Cigalino G, Caroli A, Pagnacco G (2007). Effect of k-casein polymorphism on milk composition in the orobica goat. J. Dairy Sci., 90(4): 1962-1966. https://doi.org/10.3168/jds.2006-508
Fan X, Gao S, Fu L, Qiu L, Miao Y (2020). Polymorphism and molecular characteristics of the CSN1S2 gene in river and swamp buffalo. Arch. Anim. Breed., 63(2): 345-354. https://doi.org/10.5194/aab-63-345-2020
Fazilah NF, Ariff AB, Khayat ME, Rios-Solis L, Halim M (2018). Influence of probiotics, prebiotics, synbiotics and bioactive phytochemicals on the formulation of functional yogurt. J. Funct. Foods, 48: 387-399. https://doi.org/10.1016/j.jff.2018.07.039
Gautam L, Waiz HA, Nagda RK (2020). Significance of environmental influences on average daily milk traits of sirohi goats in their native tract. Indian J. Anim. Res., 54(6): 781-785. https://doi.org/10.18805/ijar.B-3834
Gipson TA (2019). Special issue recent advances in breeding and genetics for dairy goats. Asian-Australas. J. Anim. Sci., 32(8): 1275. https://doi.org/10.5713/ajas.19.0381
Gupta SC, Kumar D, Pandey A, Malik G, Gupta N (2009). New k-casein alleles in Jakhrana goat affecting milk processing properties. Food Biotechnol., 23: 1-14. https://doi.org/10.1080/08905430802672079
Hartl DL, and Clark AG (1997). Principle of population genetic. Sinauer Associates.
Ishag IA, Abdalla SA, Ahmed MA (2012). Factors affecting milk production traits of saanen goat raised under Sudan semi arid conditions. J. Anim. Feed Res., 1(5): 435-438.
Idowu ST, Adewumi OO. (2017). Genetic and non-genetic factors affecting yield and milk composition in goats. J Adv Dairy Res., 5(2). https://doi.org/10.4172/2329-888X.1000175
Jemmali B, Gara AB, Selmi H, Ammari Z, Bouheni C, Ben LM, Hammami M, Amraoui M, Kamoun M, Rouissi H, Rekik B (2013). Kappa casein gene polymorphism in local Tunisian goats. Pak. J. Biol. Sci., 16(24): 2031-2035. https://doi.org/10.3923/pjbs.2013.2031.2035
Kalyankar SD, CDK, Patil AM (2016). Goat milk. In: (ed. B.P. Caballero). Encyclopedia of food and health (M. Finglas and F. Toldrá eds.). Elsevier Ltd. https://doi.org/10.1016/B978-0-12-384947-2.00358-5
Kiplagat SK, Agaba M, Kosgey IS, Okeyo M, Indetie D, Hanotte O, Limo MK (2010). Genetic polymorphism of kappacasein gene in indigenous Eastern Africa goat populations. Int. J. Genet. Mol. Biol., 2: 1-5.
Kumar A, Rout PK, Mandal A, Roy R (2009). Kappa-casein gene polymorphism in Indian goats. Indian J. Biotechnol., 8: 214-217. http://hdl.handle.net/123456789/3893
Lérias JR, Hernández-Castellano LE, Suárez-Trujillo A, Castro N, Pourlis A, Almeida AM (2014). The mammary gland in small ruminants: Major morphological and functional events underlying milk production. A review. J. Dairy Res., 81: 304-318. https://doi.org/10.1017/S0022029914000235
Mucha S, Mrode R, Coffey M, Kizilaslan M, Desire S, Conington J (2018). Genome-wide association study of conformation and milk yield in mixed-breed dairy goats. J. Dairy Sci., 101(3): 2213-2225. https://doi.org/10.3168/jds.2017-12919
Nei M, Kumar S (2000). Moleculear evolution and phylogenetics. Oxford University.
Patel SB, Pande AM, Rank DN, Arya JS, Jacob N (2011). Kappa casein gene polymorphism in Zalawadi goats. Indian J. Biotechnol., 10: 235-237.
Pazzola M, Dettori ML, Pira E, Noce A, Paschino P, Vacca GM (2014). Effect of polymorphisms at the casein gene cluster on milk renneting properties of the Sarda goat. Small Ruminant Res., 117(2-3): 124-130. https://doi.org/10.1016/j.smallrumres.2013.12.004
Rehman S, Feng T, Wu S, Luo X, Lei A, Luobu B, Hassan FU, Liu Q (2021). Comparative genomics, evolutionary and gene regulatory regions analysis of casein gene family in bubalus bubalis. Front. Genet., 12: 408. https://doi.org/10.3389/fgene.2021.662609
Salva S, Nuñez M, Villena J, Ramón A, Font G, Alvarez S (2011). Development of a fermented goats’ milk containing Lactobacillus rhamnosus: In vivo study of health benefits. J. Sci. Food Agric., 91(13): 2355-2362. https://doi.org/10.1002/jsfa.4467
Šlyžius E, Šlyžienė B, Lindžiūtė V (2017). Factors affecting goat milk fat yield. Zem. Ukio Moksl, 24: 91–100.30. https://doi.org/10.6001/zemesukiomokslai.v24i3.3555
Singh P, Singh MK, Rout PK, Dige MS (2018). Association of the CSN1S1 gene polymorphism with milk composition traits in Jamunapari goat. Indian J. Anim. Res., 52(8): 1107-1112. https://doi.org/10.18805/ijar.B-3176
Vacca GM, Stocco G, Dettori ML, Pira E, Bittante G, Pazzola M (2018). Milk yield, quality, and coagulation properties of 6 breeds of goats: Environmental and individual variability. J. Dairy Sci., 101(8): 7236-7247. https://doi.org/10.3168/jds.2017-14111
Volkandari SD, Indriawati, Margawati ET (2017). Genetic polymorphism of kappa-casein gene in Friesian Holstein: A basic selection of dairy cattle superiority. J. Indones. Trop. Anim. Agric., 42(4): 213-219. https://doi.org/10.14710/jitaa.42.4.213-219
Wardani FE, Palayukan J, Furqon A, Suyadi S, Susilorini TE (2022). Polymorphism of Β-Lactoglobulin (Β-Lg) gene and its association with milk yield and milk composition on Senduro goats. Iran. J. Appl. Anim. Sci., 12(1): 111-118.
Zamuner F, DiGiacomo K, Cameron AWN, Leury BJ (2020). Effects of month of kidding, parity number, and litter size on milk yield of commercial dairy goats in Australia. J. Dairy Sci., 103(1): 954-964. https://doi.org/10.3168/jds.2019-17051
To share on other social networks, click on any share button. What are these?






