Molecular Identification of Amylolytic Bacteria Isolated from Digestive Tract of Milkfish (Chanos chanos Forskal) based on 16S rRNA Gene Sequences
Molecular Identification of Amylolytic Bacteria Isolated from Digestive Tract of Milkfish (Chanos chanos Forskal) based on 16S rRNA Gene Sequences
Ummul Firmani1,3, Rahmi Nurdiani2, Arning Wilujeng Ekawati2 and
Happy Nursyam2*
1Doctoral Program, Faculty of Fisheries and Marine Science, Brawijaya University, Malang 65145, East Java, Indonesia
2>Faculty of Fisheries and Marine Science, Brawijaya University, Malang 65145, East Java, Indonesia
3Department of Aquaculture, Faculty of Agriculture, Muhammadiyah University of Gresik, Gresik 61121, East Java, Indonesia
ABSTRACT
Probiotic bacteria play an important role in fish growth and health in aquaculture, especially in fish rearing in inland waters. This study aimed to identify bacteria from the digestive tract of milkfish (Chanos chanos Forskal) and investigate their potential as probiotic candidates. Bacteria were isolated from the digestive tract of milkfish and identified using PCR technique and 16S rRNA gene sequencing. The probiotic potency was determined using amylolytic assay, synergistic activity, hemolytic activity, and antagonistic activity. The results showed that there were several types of bacteria found in the digestive tract of milkfish. One of these bacteria could produce an amylase enzyme with an amylolytic index of 5.16, identified as Bacillus paramycoides. This bacteria also synergized with other bacteria and did not have hemolytic activity on blood agar media. The results of the antagonistic test based on the well-diffused method against Aeromonas hydrophila showed that B. paramycoides did not produce an inhibition zone around the bacterial wells. The molecular identification found that the bacterial species was B. paramycoides B2.1. These results suggested that Bacillus paramycoides B2.1, which is found in the digestive tract of milkfish, can be used as a probiotic candidate for fish feed indicated by several probiotic tests that have been carried out.
Article Information
Received 16 February 2022
Revised 18 May 2022
Accepted 25 June 2022
Available online 13 October 2022
(early access)
Published 30 October 2023
Authors’ Contribution
UF designed the study and wrote the manuscript. RN performed statistical analysis. AWE interpreted the results. HN reviewed the manuscript. All authors read and approved the final manuscript.
Key words
Amylolytic, Antagonistic, Hemolytic, Milkfish, Synergistic
DOI: https://dx.doi.org/10.17582/journal.pjz/20220216070257
* Corresponding author: [email protected]
030-9923/2023/0006-2941 $ 9.00/0
Copyright 2023 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
INTRODUCTION
Milkfish (Chanos chanos Forskal) is categorized as herbivorous fish species (Djumanto et al., 2017). Milkfish are euryhaline with habitats in freshwater lakes and hypersaline lagoons (Chang et al., 2018). In their natural habitat, the larval stage of milkfish eats phytoplankton, such as chlorella, isochrysis and tetraselmis. The juvenile and adult stage of milkfish eats aquatic plants such as Klekap, complex cyanobacteria, diatoms and associated invertebrates and Bryophyta, consisting of filamentous green algae (Yap et al., 2007). High amylase activity can be found in the intestine, pancreas, pyloric caeca and liver of milkfish (Chiu and Benitez, 1981). Protease and lipase activity is high in the pyloric caeca, intestine, pancreas and esophagus (Benitez and Tiro, 1982; Borlongan, 1990).
The gastrointestinal bacterial flora of fishes can produce extracellular enzymes such as proteolytic, amylolytic, cellulolytic, lipolytic, and chitinolytic enzymes. These enzymes are involved in the digestion of proteins, carbohydrates, cellulose, lipids and chitin in the host (Bairagi et al., 2002; Ray et al., 2012). It also promotes the nutritional benefits of cultivable fish (Dutta et al., 2015). Exploring starch-degrading bacteria or amylolytic bacteria from milkfish is important to developing fish feed technology. The amylolytic bacteria can help accelerate the process of food decomposition in the fish’s body. Bacteria with the ability to produce high amylase enzymes can be used as probiotic bacteria on fish feed (Sahoo et al., 2015). Several criteria must be met so that amylolytic bacteria can be used as probiotics, including the ability to produce amylase enzymes, not hemolytic, antagonistic abilities against pathogenic bacteria, and work synergistically with other beneficial bacteria (Sahoo et al., 2015). Bermudez-Brito et al. (2012) described several mechanisms of probiotics, such as increasing the epithelial barrier, increasing adhesion to the intestinal mucosa, inhibiting the adhesion of pathogens simultaneously, competing with pathogenic microorganisms and producing anti-microbial substances and modulating the immune system.
The potential probiotic bacteria in the digestive tract of fish can be identified using the 16S rRNA gene sequence. The phylogenetic relationship between all bacterial species can be determined using this method (Khan et al., 2021). The sequence of the 16Sr RNA gene has been determined for a wide variety of bacterial species, including strains. Another advantage of 16S rRNA analysis in bacteria identification is the high accuracy, efficacy, and speed of the method (Akihary and Kolondam, 2020). Therefore, this study aimed to identify and investigate the potential probiotic bacteria isolated from the milkfish (Chanos chanos Forskal) intestine using 16S rRNA gene sequences.
Materials and methods
Isolation and characterization of bacteria
Bacteria were isolated from the digestive tract of milkfish (Chanos chanos Forskal) taken from brackish water ponds in Ujungpangkah District, Gresik, with a weight of about 65 g. The intestines were removed from the milkfish body aseptically using a sectio set. The intestines were gently excised and cut open with a pair of sterile scissors. Gut contents were removed by scraping. Then homogenates were mixed in 9 mL distilled water and diluted series until 10-4. 1 mL dilution was cultured using the spread plate method in nutrient agar and incubated at 37°C for 48 h in an incubator. To obtain a pure culture, the colonies with different morphological were streaked separately on nutrient agar tubes. Bacterial cell characteristics were tested to determine cell shape, gram properties, and bacterial motility. The biochemical or physiological characteristics of bacteria were analyzed using Microbact™ Identification Kits (OXOID) 24E (12A+12B) (Thermo Fisher Scientific Inc., US) to identify the reaction of bacterial cells to several types of sugar.
Amilolytic activity
Amilolytic activity test was conducted using Starch agar to determine the ability of bacteria to produce amylase. The method of enzymatic activity was based on Teather and Wood (1982). Enzymatic activity was observed from a bacterial colony’s inhibition zone/clear zone. The greater the clear zone index value, the greater the enzyme produced by bacteria. The enzymatic degradation power was classified based on the clear zone index value with the criteria for low, medium and high categories, respectively, where the Amilolytic index (AI) value 1, the AI value was 1-2 and the AI value was 2 (Choi et al., 2005). According to Kasana (2008), amylolytic index (AI) was obtained using the formula:
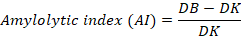
Where; AI is Amylolytic index (mm); DB is Clear zone diameter (mm), and DK is Colony diameter (mm).
Synergistic activity
The synergism test was carried out to determine whether each isolate works synergistically with the other. The bacteria tested for synergism are selected from the enzymatic activity test stages, including cellulolytic, amylolytic and proteolytic. The synergism test was carried out based on Silitonga et al. (2013). Bacterial isolates were grown in petri dish containing NA media. One petri dish contains 2-4 isolates grown by rubbing against each other using a streak plate and incubated for 24 h at room temperature ± 35oC. The formation of an inhibition zone on the touching scratches indicated that the two isolates could not work synergistically or inhibit each other.
Hemolytic activity
Hemolytic test was performed using Blood agar media (Argyri et al., 2013). Bacterial isolates were inoculated in Petri dishes containing Blood agar media by streak plate method and incubated at 37oC for 48 h. Bacterial strains that did not form a clear zone around the colony were declared non-hemolytic, whereas strains with a clear zone had the hemolytic ability.
Antagonistic activity
Antagonistic activity between probiotic candidate and pathogenic Aeromonas hydrophila using well diffusion method (Schillinger and Lucke, 1989). Each candidate probiotic and pathogenic bacteria were isolated and cultured in 30 mL Trypticase Soybean Broth (TSB) media and incubated at room temperature for 24 h. Next, isolates of pathogenic bacteria were cultured in Trypticase Soybean Agar (TSA) media using the pour plate method as much as 108 CFU/ml and incubated at 37 oC for 24 h. On the surface of the agar media that has been overgrown with Aeromonas hydrophyla, holes/wells with a diameter of 6 mm were made and filled with 30 µL of suspension of probiotic bacteria isolates (106 CFU/ml). Then, the bacteria were incubated in the incubator at 37oC for 24-48 h and observed inhibition zones’ formations.
Molecular identification and phylogenetic tree
For molecular identification the isolated strain was identified based on 16s rRNA sequence analyses. Total DNA was extracted using NEXprep™ Cell/ Tissue DNA Mini Kit (NEX Diagnostics, Korea). The DNA extraction protocol was conducted according to the manufacturer protocol of the kits. Polymerase chain reaction (PCR) amplifies the 16S rRNA with the primer 16 S universal 1492R 5’ TACGGYTACCTTGTTACGACTT 3’, 27F 5’ AGAGTTTGATCMTGGCTCAG 3’. The PCR products were sequenced by 1stBASE Laboratories Sdn Bhd, Malaysia. The neighbour-joining method was used to construct phylogenetic trees using MEGA-X software version 10.2.5 (Penn State University, US) to determine the most likely bacterial strain phylogeny.
Results
Morphology of bacteria
The characteristic of bacteria colonies from milkfish gut was shown in Figure 1A. The results showed that colors, sizes, and shapes varied among the colonies on the nutrient agar plate. Each colony was purified in a separate tube and observed for the characteristics of bacterial cells. One of the colonies was further analyzed. Colonies were pink, round in shape with jagged edges, an orange colony circle in the middle, and the size of the colony was 4.23 cm in diameter (Fig. 1A). The purified bacteria were categorized as gram-negative bacteria with the basil in shape, motile, positive catalase, spores, and positive gelatine (Supplementary Table I).
Amylolytic activity
Screening amylolytic activity of bacterial isolates from the digestive tract of freshwater milkfish showed that the clear zone width was 12.27 mm, and the bacterial colony width was 2 mm (Fig. 1B). Thus, the average clear zone index on amylolytic activity was 5.16.
Synergistic activity
The bacterial isolates B2.1 gave positive synergistic activity tests indicated by the absence of a clear zone or inhibition zone around the bacterial colonies that were scratched together (Fig. 2). The synergistic activity of bacteria is important when making a consortium bacterial culture that will be used as a probiotic.
Hemolytic activity
Figure 3 showes that the bacterial isolate B2.1 had gamma hemolytic activity, which suggested that it did not have red blood cell lysing activity.
Antagonistic activity
Figure 4 shows the antagonistic test of B. paramycoides B2.1 against pathogens Aeromonas hydrophila. There was no clear zone around the well containing B. paramycoides B2. 1 culture, indicating that B. paramycoides B2.1 has no antagonistic activity with Aeromonas hydrophila. It is possible that B. paramycoides strain B21 lacked antagonistic activity against A. hydrophila because its antimicrobial activity was weak.
Phylogenetic tree
Based on the Basic Local Alignment Search Tool (BLAST) 16S rRNA sequence, of strain B2.1 had a maximum similarity of 98.26% to Bacillus paramycoides (Accession number: NR_157734.1), which was supported by phylogenetic analysis (Fig. 5). Therefore, it was identified as Bacillus paramycoides B2.1.
Discussion
The herbivorous fish can produce the cellulase enzyme which is mediated by specific microorganisms (Li et al., 2008; Saha et al., 2006). The presence of high amylolytic activity in bacteria isolated from the digestive tract of milkfish is since milkfish are herbivores, so most of the composition of the food comes from plants. Amylase is a digestive enzyme that contributes in the breakdown of carbohydrates by hydrolyzing the bonds between sugar molecules in polysaccharides. It is important to digest starch into sugars to make available energy sources for the body (Bhilave et al., 2014).
Istifadah et al. (2014) stated that an isolate was said to be compatible if there was no zone of inhibition at the meeting area of the two isolates, and it was said to be incompatible if there was an inhibition zone at the meeting area of the two isolates. The synergism test revealed that B2.1 isolates had the potential to be developed as probiotic bacteria. The ability to work synergistically is important in a bacterial consortium where bacteria grown in the same medium will complement each other’s characteristics. This characteristic is necessary for the culture of probiotic bacteria, where different types of bacteria work synergistically rather than compete.
The haemolytic activity assay is considered to be an important probiotic screening process. Hemolysin is a prevalent virulence factor, which frequently causes anemia and edema in the host, and hence, haemolytic strains should not be used as feed additives (Ouwehand et al., 2005). Therefore, the non-haemolytic strains would be preferable for probiotic use (Nandi et al., 2017). The present study confirmed that the isolated Bacillus strains did not show any haemolytic activity, and hence it can be used with food ingredients for better health. Similarly, Ramesh et al. (2015) have confirmed that Bacillus spp. showed non-haemolytic activity. Deng et al. (2021) concluded that no hemolysis and cytotoxicity was observed, and no presence of toxin genes was positively detected in 20 Bacillus spp. This indicates the safety of using these Bacillus isolates as potential probiotics.
Cultured supernatants on agar media containing pathogenic bacteria using paper disks is another method for detecting the existence of antagonistic or antibacterial activity. Since the well approach produced minimal levels of antimicrobial due to the tiny number of bacteria in the wells, antagonistic activity was not detected in the agar media. Dharmaraj et al. (2020) revelaed that B. paramycoides had antagonistic activity against the bacteria V. parahaemolyticus, Salmonella sp., Enterobacter sp. and Micrococcus sp. According to the findings of this study, isolate B2.1 was identified as Bacillus paramycoides, a non-hemolytic with the ability to work synergistically with other bacteria from the milkfish digestive tract.
Conclusion
Bacillus paramycoides B2.1 was identified as a bacterial species isolated from the digestive tract of milkfish. Bacillus paramycoides B2.1 is a starch-degrading bacteria that can work synergistically with three other bacteria species found in the digestive tract of milkfish, however, it lacks hemolytic activity. These results suggested that Bacillus paramycoides B2.1 can be considered a probiotic candidate for fish feed.
Acknowledgments
This study was funded by the Ministry of Education and Culture of the Republic of Indonesia through Domestic Postgraduate Education Scholarships.
There is supplementary material associated with this article. Access the material online at: https://dx.doi.org/10.17582/journal.pjz/20220216070257
Statement conflict of interest
The authors have declared no conflict of interest.
References
Akihary, C.V. and Kolondam, B.J., 2020. Utilization of the 16S RRNa gene as a bacterial identification device for research in Indonesia. Pharmacon, 9: 16–22. https://doi.org/10.35799/pha.9.2020.27405
Argyri, A.A., Zoumpopoulou, G., Kimon, A.G., Karatzas, K.A., Tsakalidou, E., Nychas, G.J., Panagou, E. and Tassou, C., 2013. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Fd. Microbiol., 33: 282–291. https://doi.org/10.1016/j.fm.2012.10.005
Bairagi, A., Ghosh, K., Sen, S.K. and Ray, A.K., 2002. Enzyme producing bacterial flora isolated from fish digestive tracts. Aquacult. Int., 10: 109–121. https://doi.org/10.1023/A:1021355406412
Benitez, L.V. and Tiro, L.B., 1982. Studies on the digestive proteases of the milkfish Chanos chanos. Mar. Biol., 71: 309-315. https://doi.org/10.1007/BF00397047
Bermudez-Brito, M., Plaza-Diaz, J., Muñoz-Quezada, S., Gómez-Llorente, C. and Gil, A., 2012. Probiotic mechanisms of action. Annls Nutr. Metab., 61: 160–174. https://doi.org/10.1159/000342079
Bhilave, M., Nalawade, V. and Kulkarni, J.J., 2014. Amylase activity of fingerlings of freshwater fish Labeo rohita fed on formulated feed. Int. J. Fish. aquat. Stud., 2: 53-56.
Borlongan, I.G., 1990. Studies on the digestive lipases of milkfish Chanos chanos. Aquaculture, 89: 315-325. https://doi.org/10.1016/0044-8486(90)90135-A
Chang, C.H., Huang, J.J., Yeh, C.Y., Tang, C.H., Hwang, L.Y. and Lee, T.H., 2018. Studies on the carbohydrates in the digestive tract of the milkfish Chanos chanos. Salinity effects on strategies of glycogen utilization in livers of euryhaline milkfish (Chanos chanos) under hypothermal stress. Front. Physiol., 9: 1-13. https://doi.org/10.3389/fphys.2018.00081
Chiu, Y.N. and Benitez, L.V., 1981. Enzyme production by endophytes of Brucea javanica. Mar. Biol., 61: 247-254. https://doi.org/10.1007/BF00386667
Choi, Y.W., Hodgkiss, I.J. and Hyde, K.D., 2005. Enzyme production by endophytes of Brucea javanica. J. Agric. Technol., 1: 55-66.
Deng, S., Silimon, R.L., Balakrishnan, M., Bothe, I., Juros, D., Soffar, D.B. and Baylies, M.K., 2021. The actin polymerization factor diaphanous and the actin severing protein flightless I collaborate to regulate sarcomere size. Dev. Biol., 469: 12-25. https://doi.org/10.1016/j.ydbio.2020.09.014
Dharmaraj, D., Krishnamoorthy, M., Rajendran, K., Karuppiah, K., Annamalai, J., Durairaj, K.R., Santhiyagu, P. and Ethiraj, K., 2020. Antibacterial and cytotoxicity activities of biosynthesized silver oxide (Ag2O) nanoparticles using Bacillus paramycoides. J. Drug Deliv. Sci. Technol., 61: 102111. https://doi.org/10.1016/j.jddst.2020.102111
Djumanto, D., Pranoto, B.E., Diani, V.S. and Setyobudi, E., 2017. Food and the growth of introduced milkfish, Chanos chanos (Forsskål, 1775) in Sermo Reservoir, Kulon Progo. J. Iktiol. Indones., 17: 83-100. https://doi.org/10.32491/jii.v17i1.306
Dutta, D., Banerjee, S., Mukherjee, A. and Ghosh, K., 2015. Selection and probiotic characterization of exoenzyme-producing bacteria isolated from the gut of Catla catla (Actinopterygii: Cypriniformes: Cyprinidae). Acta Ichthyol. Piscat., 45: 373–384. https://doi.org/10.3750/AIP2015.45.4.05
Istifadah, N., Melawati, A., Suryatmana, P. and Fitriatin, B.N., 2014. The effectiveness of consortium of microbial antagonists and biofertilizer to suppress damping off disease (Rhizoctonia solani) in chili. Agric. Sci. J., 4: 337-345.
Kasana, R.C., 2008. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. Microbiol., 57: 503-507. https://doi.org/10.1007/s00284-008-9276-8
Khan, M.I.R., Kamilya, D., Choudhury, T.G., Tripathy, P.S. and Rathore, G., 2021. Deciphering the probiotic potential of Bacillus amyloliquefaciens COFCAU_P1 isolated from the intestine of Labeo rohita through in vitro and genetic assessment. Probiot. Antimicrob. Proteins, 13: 1572–1584. https://doi.org/10.1007/s12602-021-09788-2
Li, H., Zheng, Z., Cong-xin, X., Bo, H., Chao-yuan, W. and Gang, H., 2008. Isolation of cellulose-producing microbes from the intestine of grass carp (Ctenopharyngodon idellus). In: Developments in environmental biology of fishes (eds. D.L.G. Noakes, A. Romero, Y. Zhao and Y. Zhou), vol 28. Springer, Dordrecht, pp. 131-135. https://doi.org/10.1007/978-90-481-3458-8_19
Nandi, A., Dan, S.K., Banerjee, G., Ghosh, P., Ghosh, K., Ringø, E. and Ray, A.K., 2017. Probiotic potential of Autochthonous bacteria isolated from the gastrointestinal tract of four freshwater teleosts. Probiot. Antimicrob. Proteins, 9: 12–21. https://doi.org/10.1007/s12602-016-9228-8
Ouwehand, A., Vankerckhoven, V., Goossens, H., Huys, G., Swings, J., Vancanneyt, M. and Lähteenmäki, A., 2005. The safety of probiotics in foods in Europe and its legislation. In: Probiotics in food safety and human health (eds. I. Goktepe, V.K. Juneja and M. Ahmedna). CRC Press, USA. pp. 405–429. https://doi.org/10.1201/9781420027570.ch18
Ramesh, D., Vinothkanna, A., Rai, A.K. and Vignes, V.S., 2015. Isolation of potential probiotic Bacillus spp. and assessment of their subcellular components to induce immune responses in Labeo rohita against Aeromonas hydrophila. Fish Shellfish Immunol., 45: 268–276. https://doi.org/10.1016/j.fsi.2015.04.018
Ray, A.K., Ghosh, K. and Ringø, E., 2012. Enzyme-producing bacteria isolated from fish gut: A review. Aquacult. Nutr., 18: 465–492. https://doi.org/10.1111/j.1365-2095.2012.00943.x
Saha, S., Roy, R.N., Sen, S.K. and Ray, A.K., 2006. Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquacult. Res., 37: 380–388. https://doi.org/10.1111/j.1365-2109.2006.01442.x
Sahoo, T.K., Jena, P.K., Nagar, N., Patel, A.K. and Seshadri, S., 2015. In vitro evaluation of probiotic properties of lactic acid bacteria from the gut of Labeo rohita and Catla catla. Probiot. Antimicrob. Prot., 7: 126–136. https://doi.org/10.1007/s12602-015-9184-8
Sahu, M.K., Swarnakumar, N.S., Sivakumar, K., Thangaradjou, T. and Kannan, L., 2008. Probiotics in aquaculture: importance and future perspectives. Indian J. Microbiol., 48: 299–308. https://doi.org/10.1007/s12088-008-0024-3
Schillinger, U. and Lücke, F.K., 1989. Antibacterial activity of Lactobacillus sake isolated from meat. Appl. environ. Microbiol., 55: 1901–1906. https://doi.org/10.1128/aem.55.8.1901-1906.1989
Silitonga, D.M., Priyani, N. and Nurwahyuni, I., 2013. Isolation and potential test of isolate phosphate solubilizing bacteria and IAA hormone-producing bacteria (indole acetic acid) against soybean (Glycine max L.) growth in yellow soil. Saintia Biol., 2: 35-41.
Teather, R.N. and Wood, P.J., 1982. Use of congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. environ. Microbiol., 43: 777-780. https://doi.org/10.1128/aem.43.4.777-780.1982
Yap, W.G., Villaluz, A.C., Soriano, M.G.G. and Santos, M.N., 2007. Milkfish production and processing technologies in the Philippines. Milkfish Project Publication Series No. 2, The WorldFish Center, Philippines. pp. 96.
To share on other social networks, click on any share button. What are these?









