Molecular Characterization and Phylogenetic Analysis of Fowl Adenoviruses Isolated from Commercial Poultry Flocks in Pakistan during 2014-15
Molecular Characterization and Phylogenetic Analysis of Fowl Adenoviruses Isolated from Commercial Poultry Flocks in Pakistan during 2014-15
Abdul Wajid1,*, Asma Basharat2, Muhammad Akbar Shahid3, Sidra Tul Muntaha1, Abdul Basit4, Tanveer Hussain5, Muhammad Farooq Tahir6, Muhammad Azhar7, Masroor Ellahi Babar1 and Shafqat Fatima Rehmani2
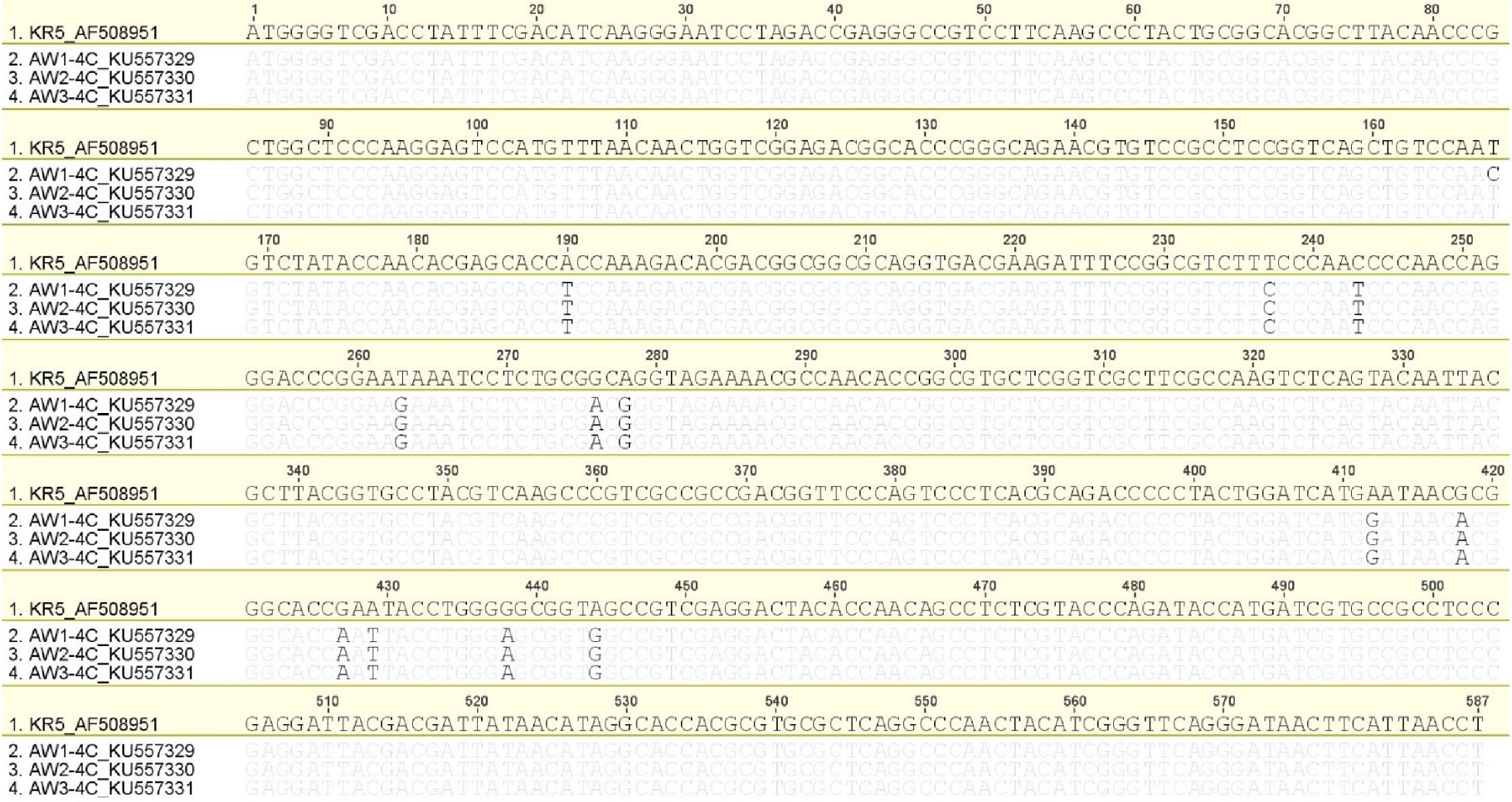
Nucleotide sequence alignment of Pakistani FAdV-4 strains/isolates with reference strain KR5 (AF508951). Differences with reference strain are highlighted.
Predicted amino acid sequence alignment of Pakistani FAdV-4 strains/isolates with reference strain KR5 (protein ID AAN77077). Differences with reference strain are highlighted.
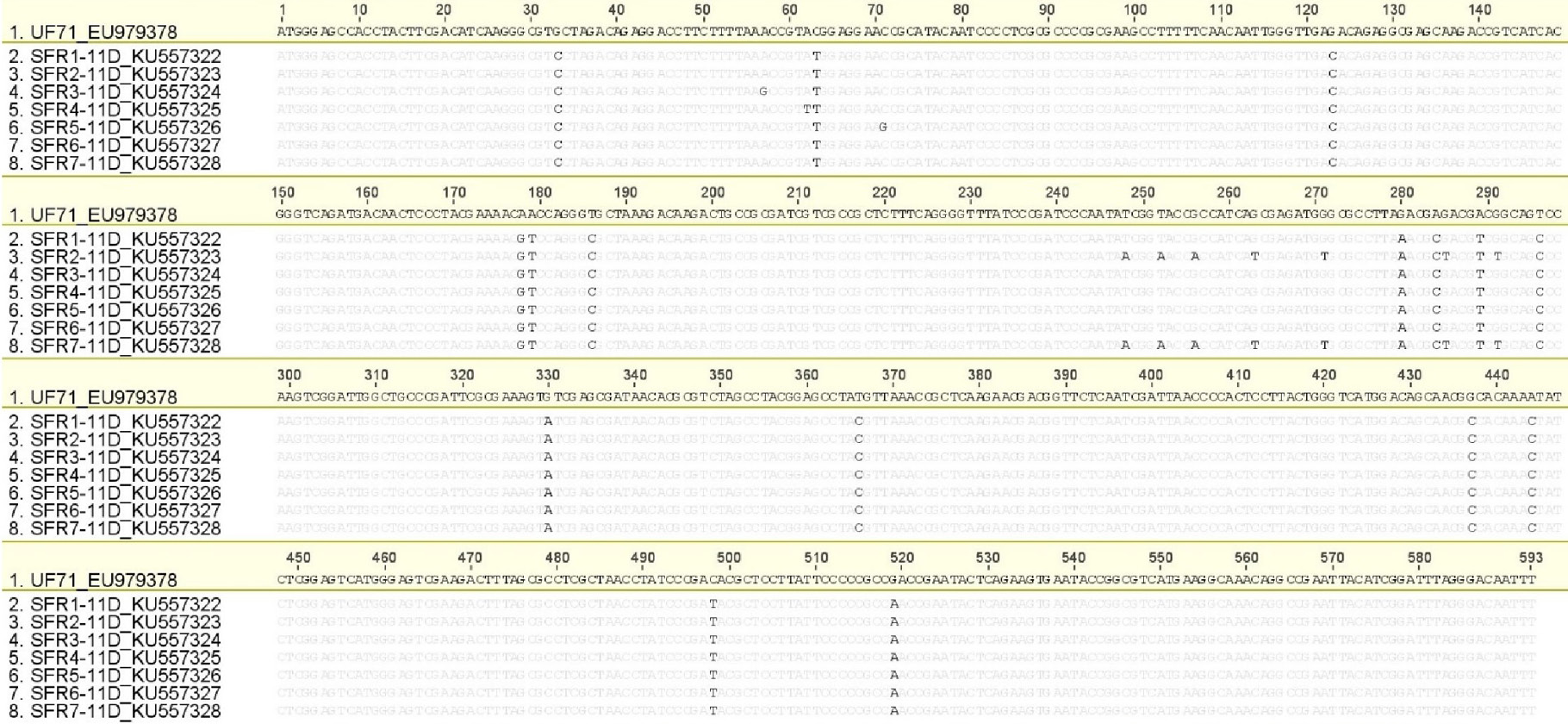
Nucleotide sequence alignment of Pakistani FAdV-11 strains/isolates with reference strain UF71 (EU979378). Differences with reference strain are highlighted.
Predicted amino acid sequence alignment of Pakistani FAdV-11 strains/isolates with reference strain KR5 (protein ID ACL68146). Differences with reference strain are highlighted.
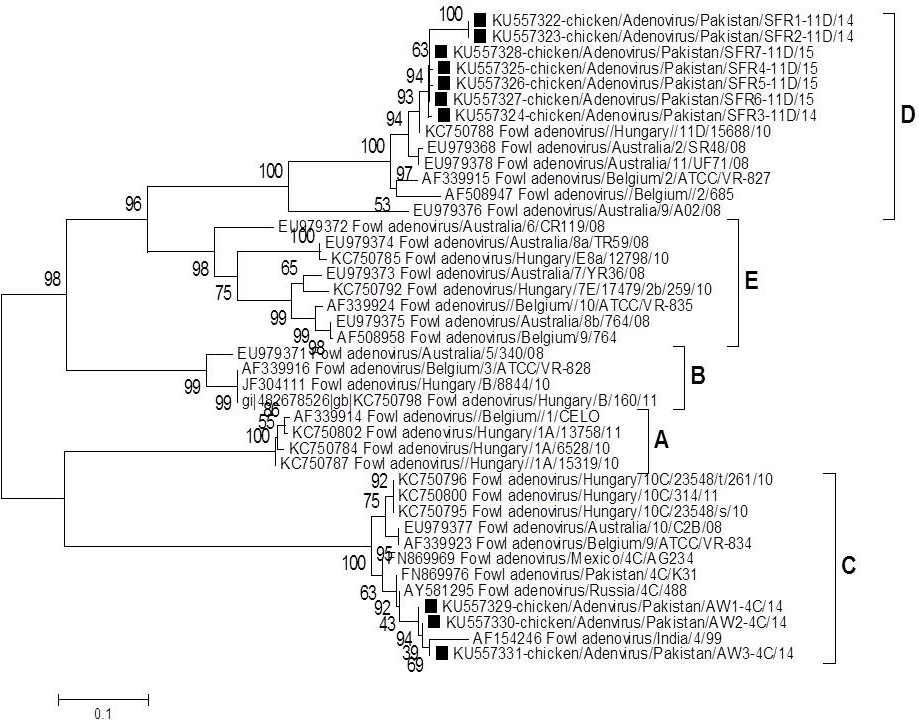
Phylogenetic analysis of the selected fowl adenoviruses (FAdVs) using nucleotide sequences encoding for the hexon gene. The data included Pakistani isolates, indicated with Black Square, and other already published FAdV strains retrieved from GenBank. FAdV strains are classified on species level e.g. species A, species B, species C, species D and species E. The evolutionary history was inferred using the Neighbor-Joining method. Numbers indicate the bootstrap values (1000 replicates). The analysis involved 42 nucleotide sequences.
Phylogenetic analysis of the selected fowl adenoviruses (FAdVs) using nucleotide sequences encoding for the hexon gene. The data included Pakistani isolates, indicated with Black Square, and other already published FAdV strains retrieved from GenBank. FAdV strains are classified on serotype level 2,3,9,11 (species D), 5 (species B), 6,7,8a,8b (species E), 1 (species A), 4,10 (species C). The evolutionary history was inferred using the Neighbor-Joining method. The analysis involved 85 nucleotide sequences.