Isolation and Complete Capsid Sequence of Enterovirus D111 from Faeces of a Child with Acute Flaccid Paralysis in Nigeria
Isolation and Complete Capsid Sequence of Enterovirus D111 from Faeces of a Child with Acute Flaccid Paralysis in Nigeria
Faleye Temitope Oluwasegun Cephas1,2*, Adewumi Moses Olubusuyi1, Olayinka Olaitan Titilola3 and Adeniji Johnson Adekunle1,3
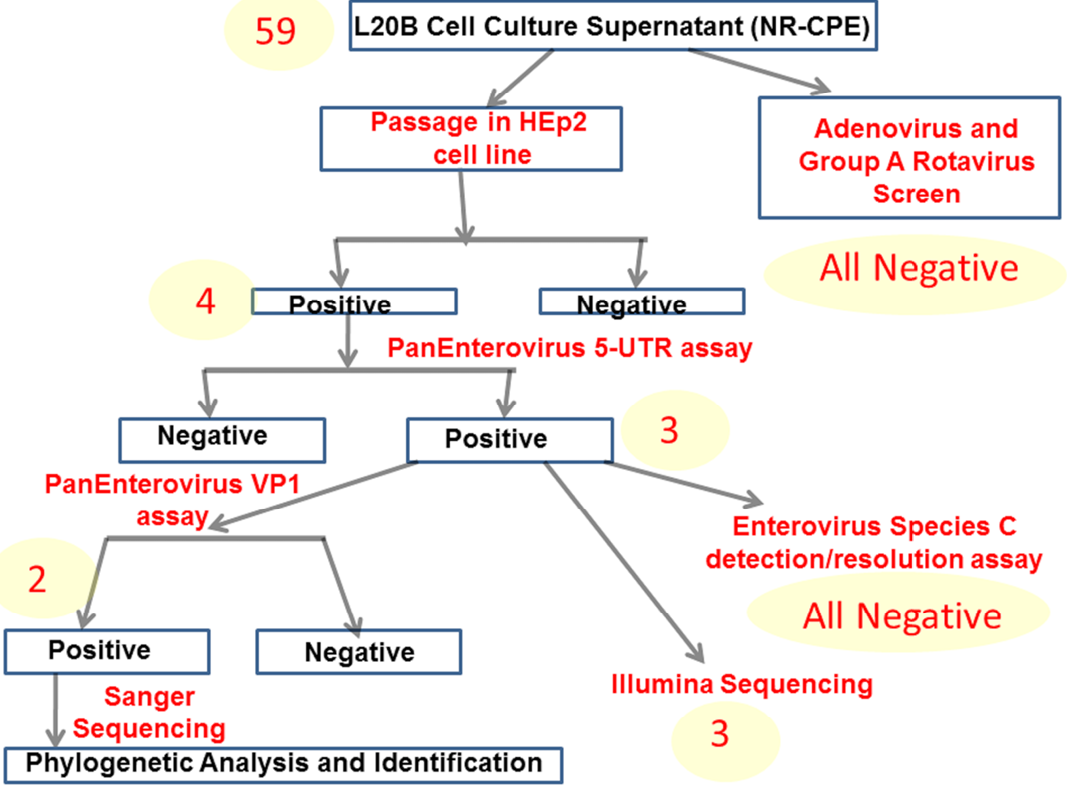
A schematic representation of the algorithm followed in this study.
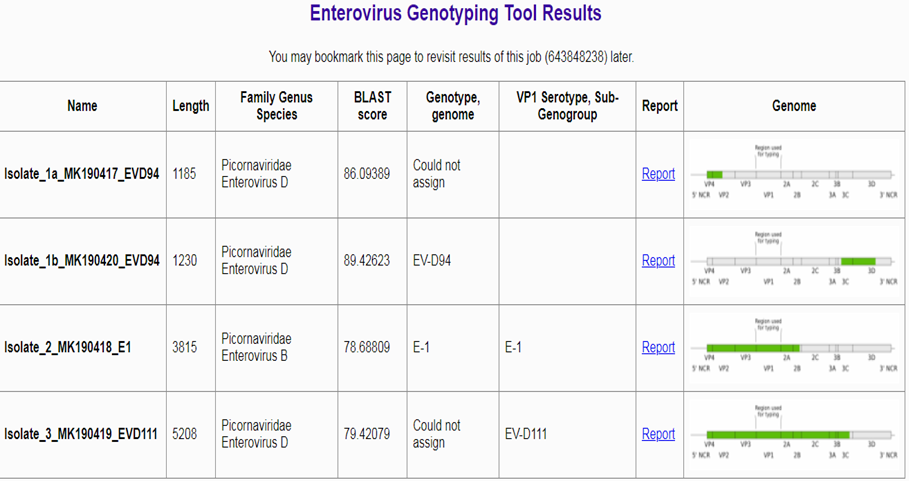
The Contigs assembled from the Illumina sequencing data generated for the three PE-5UTR positive isolates. The contigs and the genomic regions covered are displayed using the EGT.
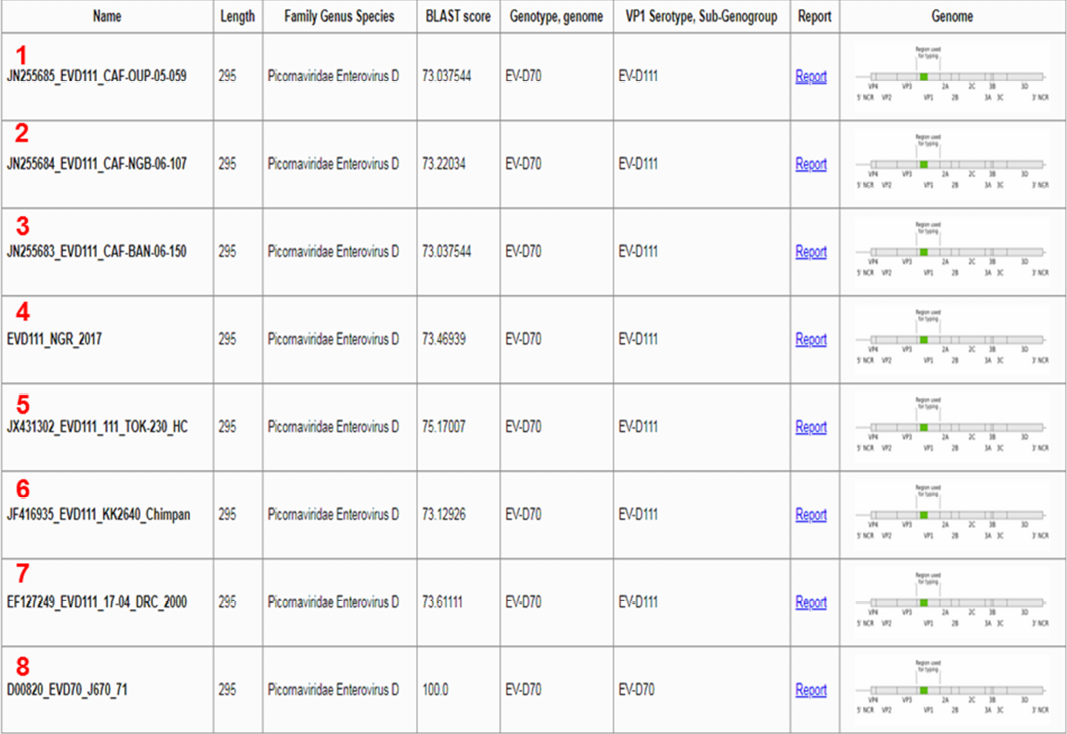
All the EV-D111 sequences in GenBank described till date alongside a reference EV-D70 and the EGT identification glitch.
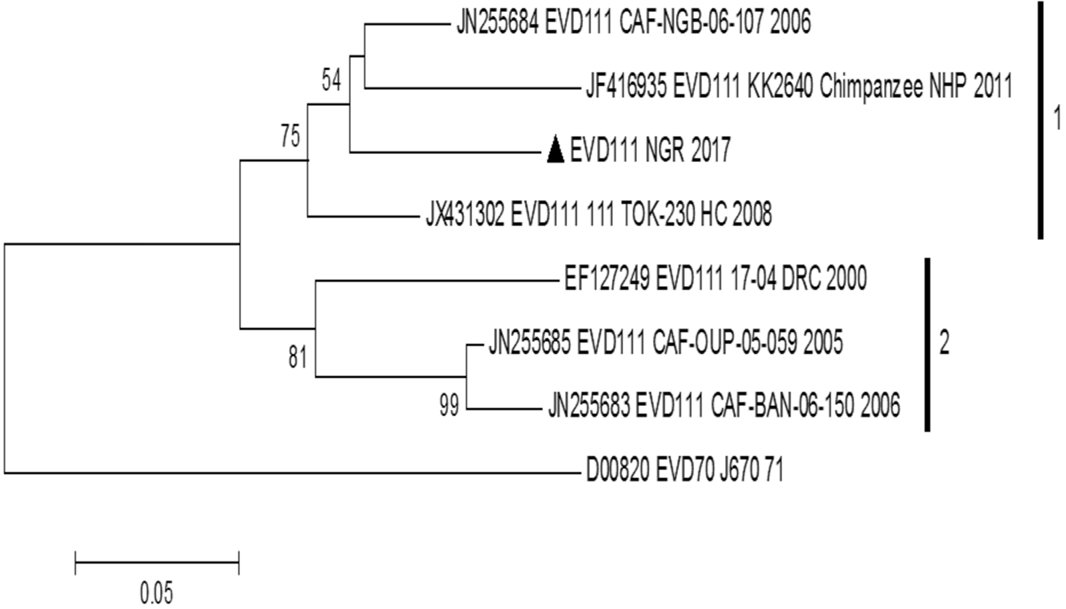
Phylogram of partial VP1 sequences of EV-D111. The phylogenetic tree is based on an alignment of partial EV-D111 VP1 sequences available in GenBank and the one described in this study. The EV-D111 described in this study is indicated with a black triangle. Bootstrap values are indicated if >50%.
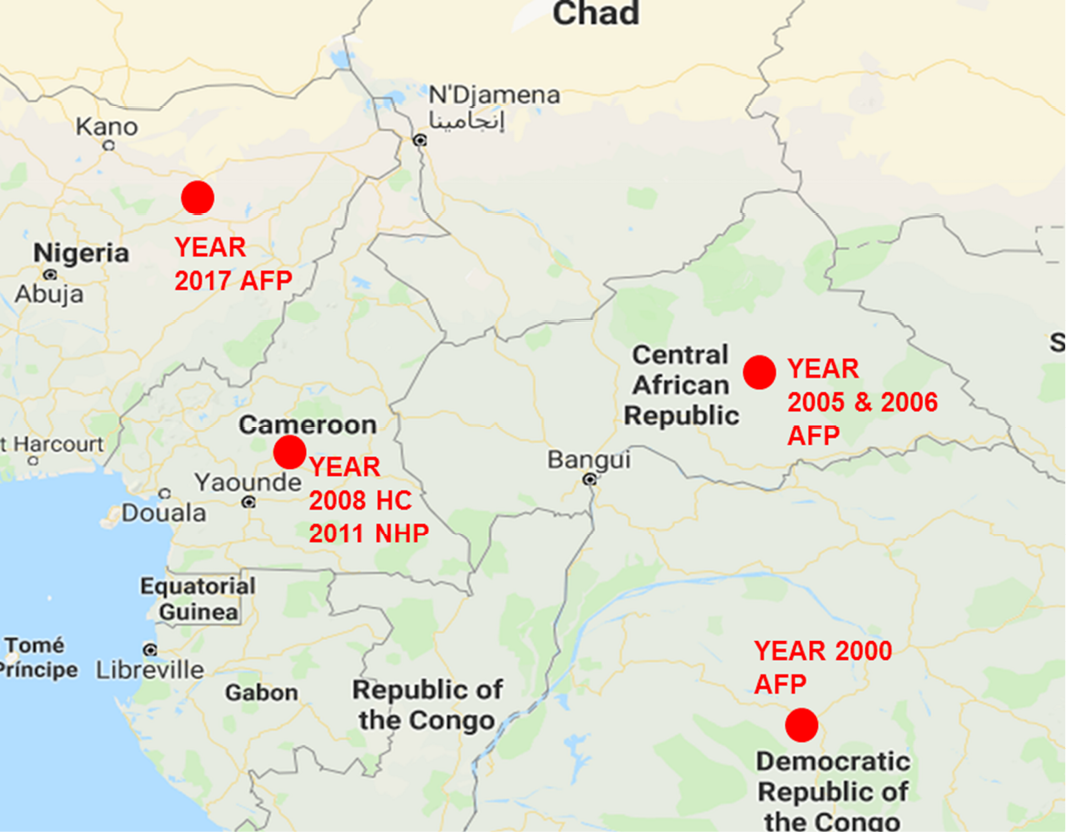
Global distribution of EV-D111 detection sources. It is crucial to note that, till date, all have been detected in sub-Saharan Africa.