Identification of Glycolysis-Related Gene Signature and Prediction on Prognosis of Colon Cancer
Qingsen Ran1,2, Manjing Li3, Jiayin Han3, Lifang Wang3, Han Wang3,4, Shaobo Liu5* and Yanping Wang1*
1Institute of Basic Research in Clinical Medicine, China Academy of Chinese Medical Science, Beijing 100700, China
2Postdoctoral Research Station, China Academy of Chinese Medical Science, Beijing 100700, China
3Institute of Chinese Materia Medica, China Academy of Chinese Medical Science, Beijing 100700, China
4Affiliated Hospital of Changchun University of Chinese Medicine, Changchun 130021, China
5The Key Laboratory of Optimal Utilization of Natural Medicine Resources, School of Pharmaceutical Sciences, Guizhou Medical University, Guiyang 550025, China
Qingsen Ran and Manjing Li contributed equally to this work.
* Corresponding author: liushaobo@gmc.edu.cn, ypwang@ircm.ac.cn
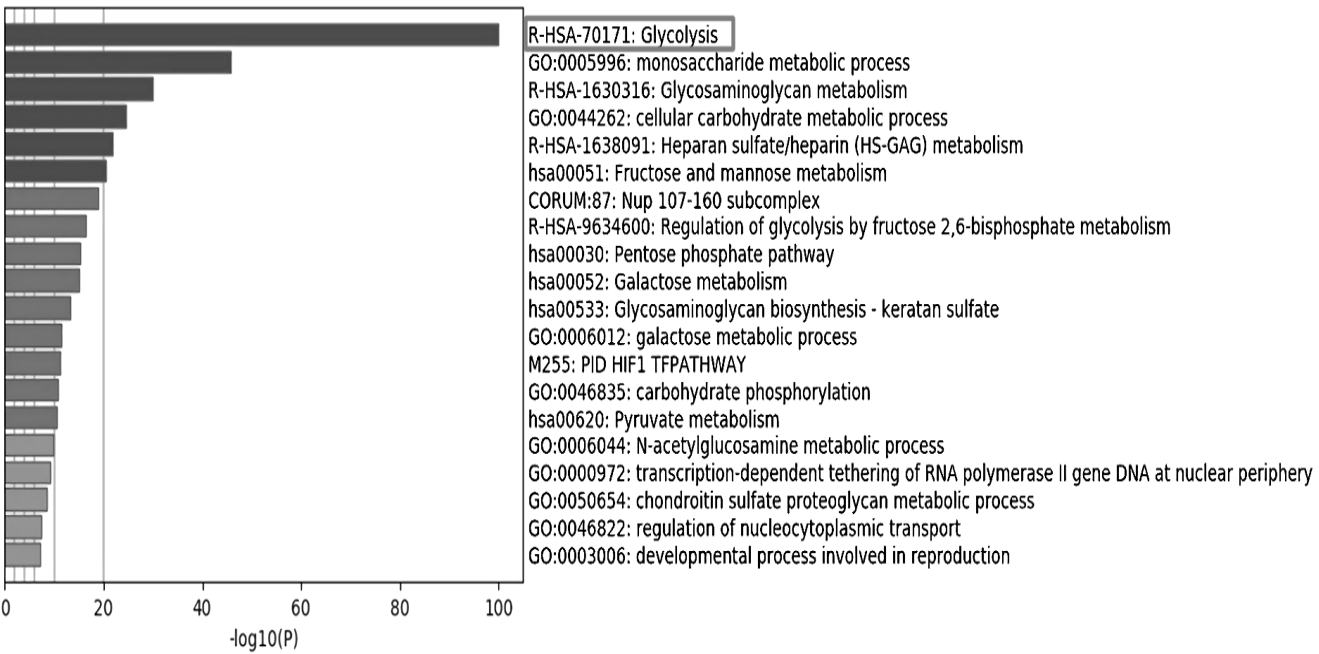
Fig. 1.
GO and KEGG pathway enrichment analysis of significant glycolysis genes based on Metascape.
Note: the significance increased from bottom to top; marked in box indicated the most significant metabolic pathway.
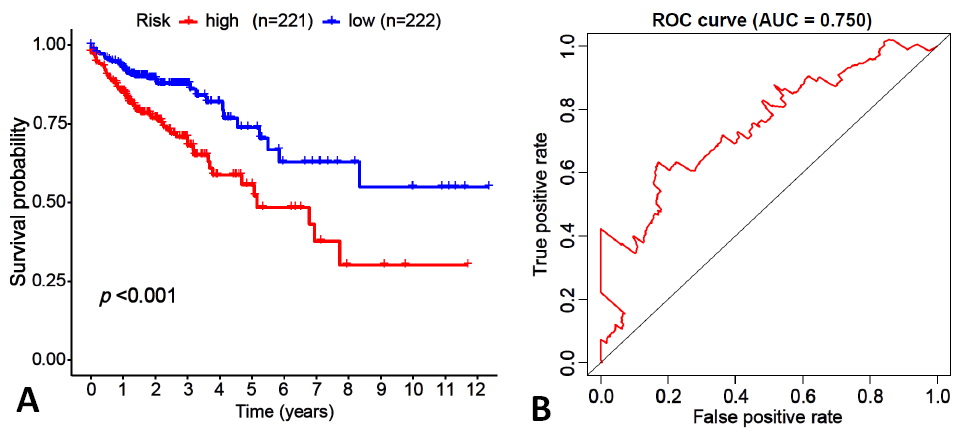
Fig. 2.
Survival curve analysis (A) and receiver operating characteristic (ROC) curve analysis (B) of glycolysis prognosis model with colon cancer patients.
Notes: A: red indicates high-risk patients (n= 221) and blue indicates low-risk patients (n= 222), and with a significant difference in survival rate (p <0.001). B: ROC curve analysis result was 0.750.
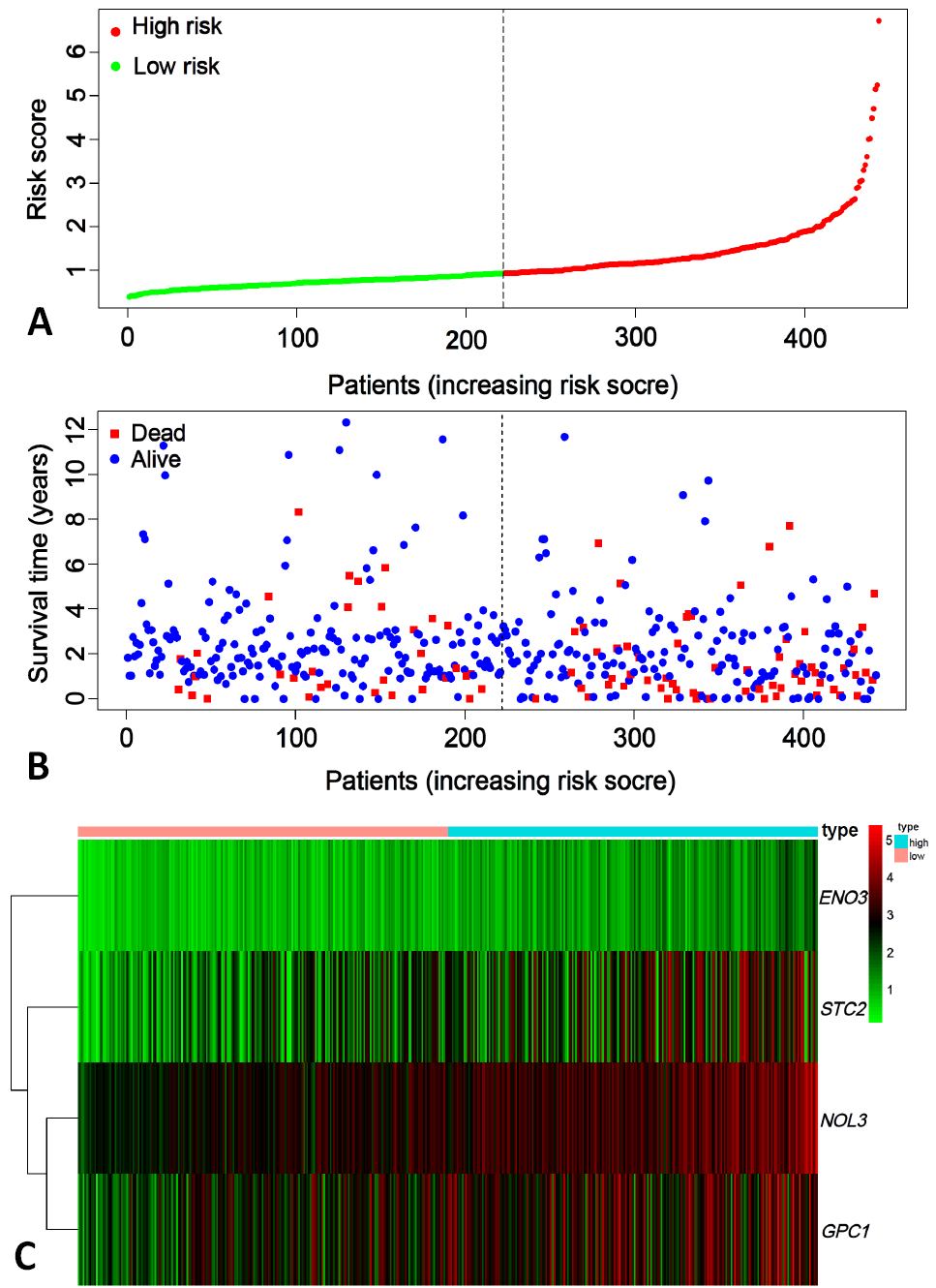
Fig. 3.
Risk map for glycolysis prognosis model in colon cancer patients.
A: risk score. B: the survival number of low risk score patients was significantly more than that of high risk score patients. C: heat map analysis. ENO3, GPC1, NOL3, STC2 genes expression in patients with high risk scores were significantly more than those with low risk scores.
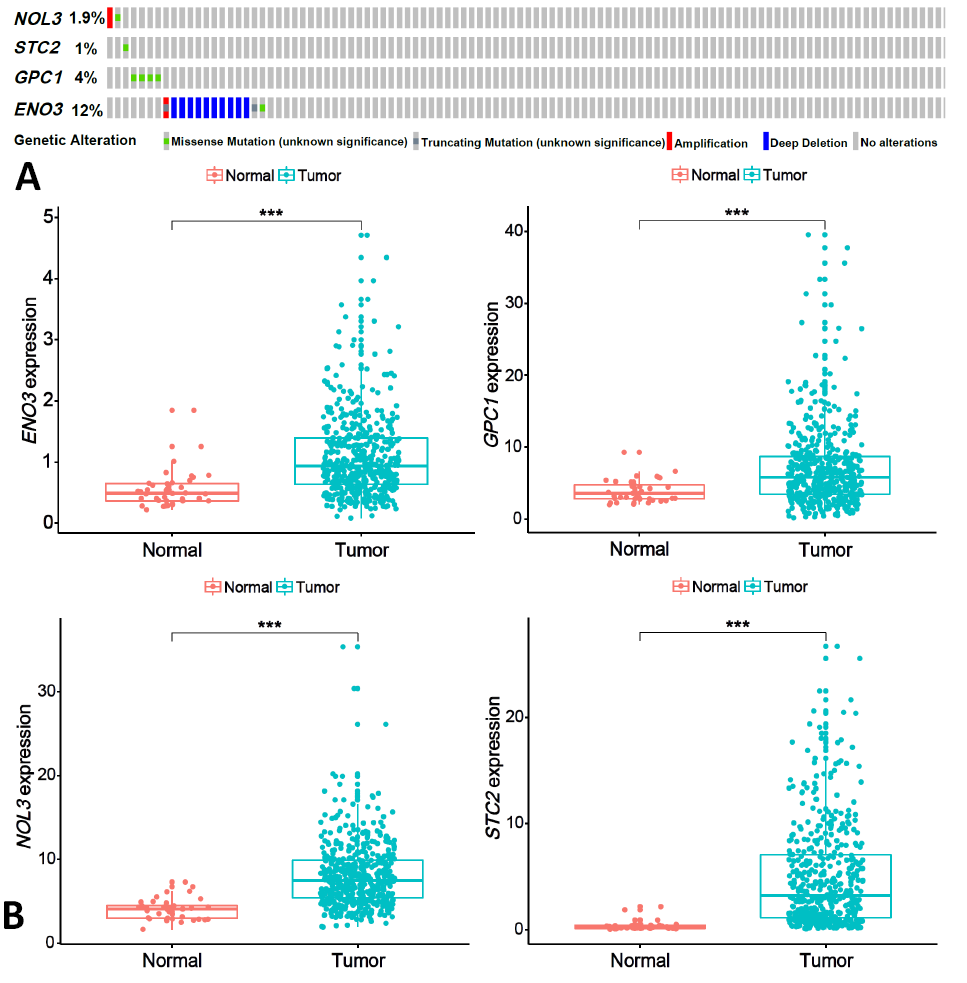
Fig. 4.
Gene mutation analysis (A) and difference analysis (B) for model samples from TCGA.
Notes: A: 105 patients samples in cBioPortal online database; NOL3: gene amplification, 1 case; missense mutation, 1 case; STC2: missense mutation, 1 case; GPC1: missense mutation, 4 cases; ENO3: deep deletion, 10 cases; truncation mutation, 1 case; missense mutation, 1 case. B: Red color: normal samples; green color: tumor samples; ***: significant difference at p <0.001.
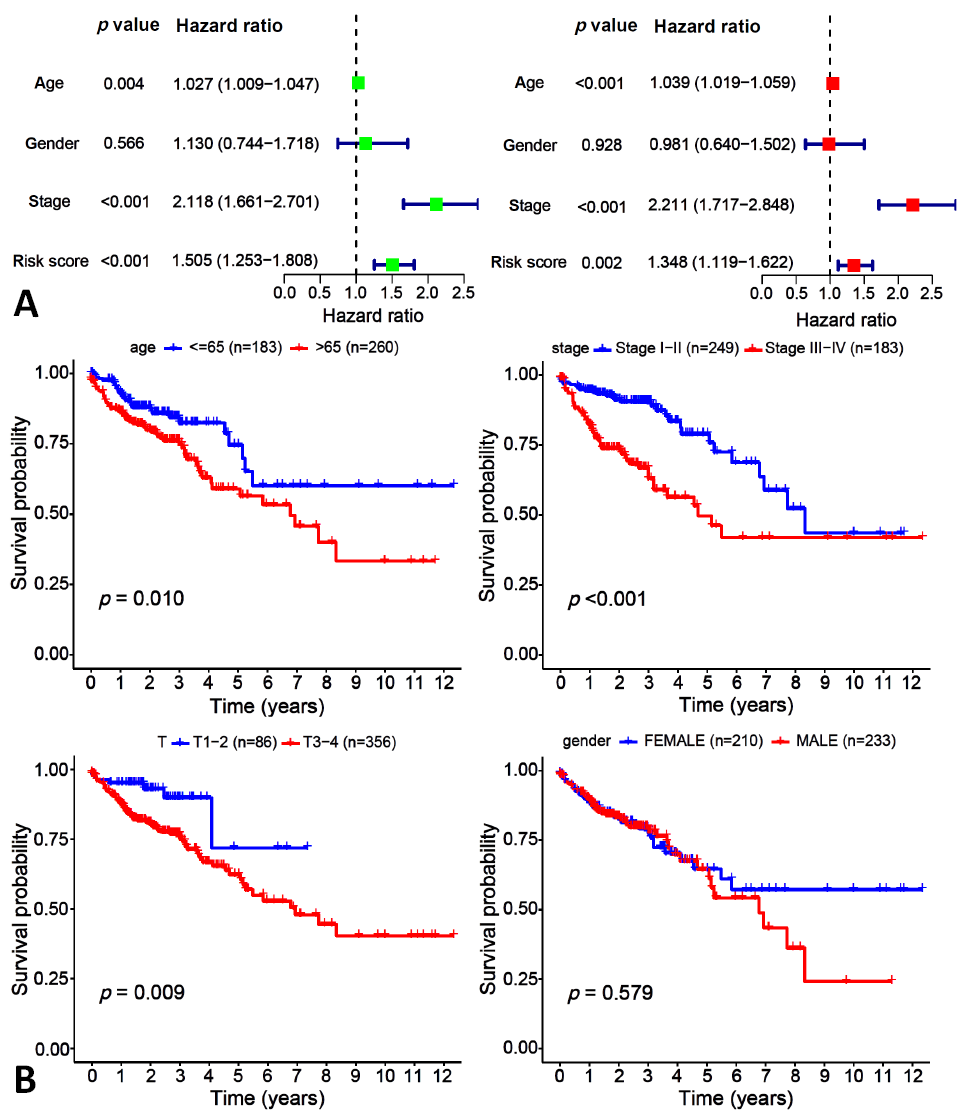
Fig. 5.
Independent prognostic analysis (A) and clinical characteristics analysis (B) for the glycolysis prognosis model. A: uniForest and multForest analysis from left to right, the stage and risk scores were statistically significant (p-value <0.05, Hazard ratio >1); B: 470 tumor samples with colon cancer based on TCGA.
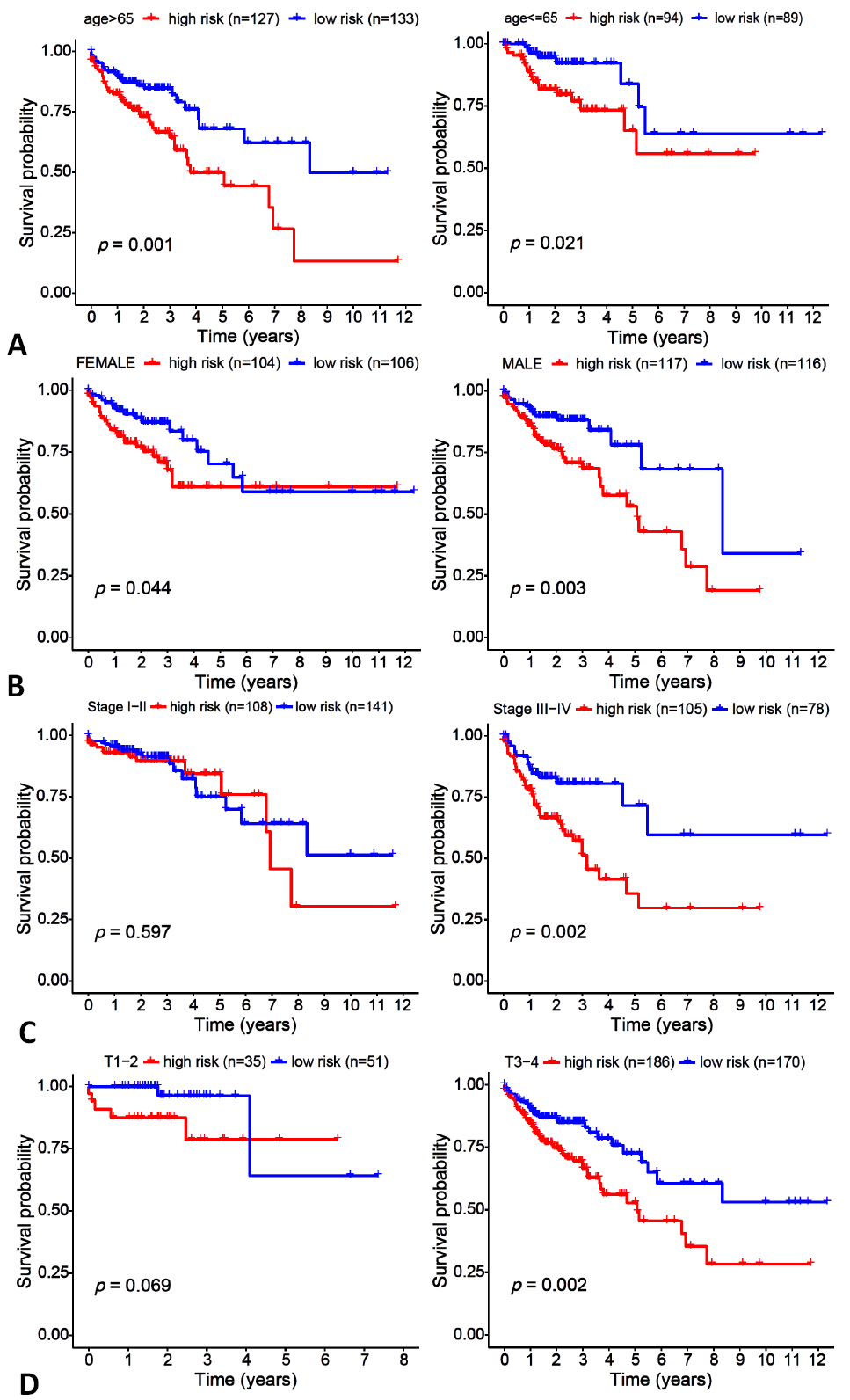
Fig. 6.
Clinical grouping verification for colon cancer patients.
A: based on age; B: based on gender; C: based on stage; D: based on T.
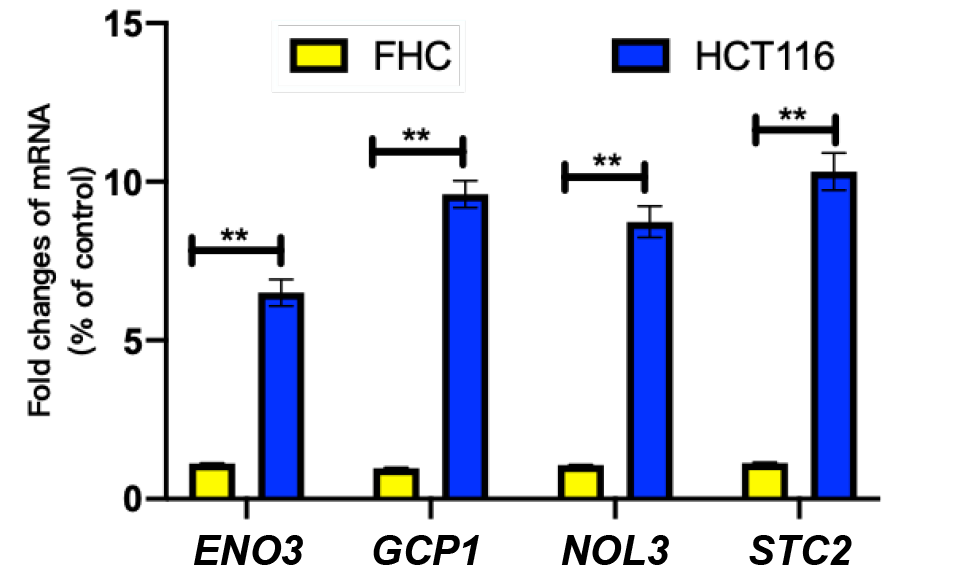
Fig. 7.
Relative expression of ENO3, GCP1, NOL3, STC2 in FHC and HCT116 cells.
FHC, human normal colorectal mucosa epithelial cells; HCT116, Human colorectal cancer cell line; Data were representative of three separate trials; ** P <0.01.