Genetic Homogeneity among Bull Sharks Carcharhinus leucas in the South China Sea
Genetic Homogeneity among Bull Sharks Carcharhinus leucas in the South China Sea
Zhaochao Deng1, Jingchen Chen1, Na Song2, Yongzhen Li3 and Zhiqiang Han1,*
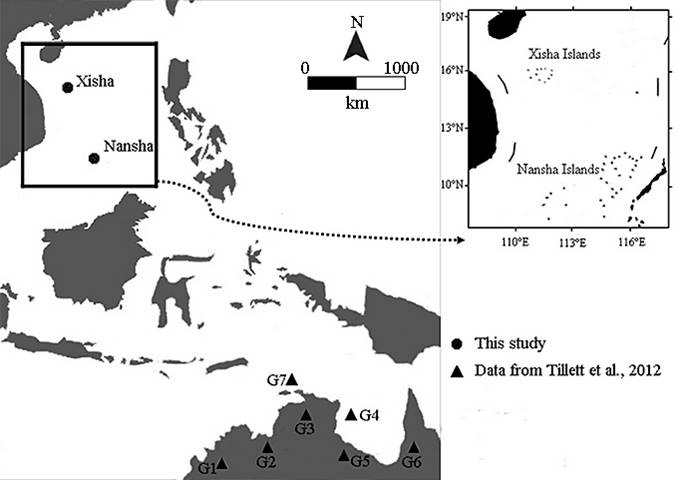
Sampling localities for this study. Triangles represent populations from Tillett et al. (2012). G1, Mitchell River, Robison River and Fitzroy River; G2, Daly River and Ord River; G3, East Alliagator River; G4, Blue Mud Bay; G5, Roper River, Towns River, Limmen River and Robinson River; G6, Wenlock River, Mission River and Mitchell River; G7, Tiwi Islands. Circles represent locations sampled in this study (Xisha and Nansha Islands).
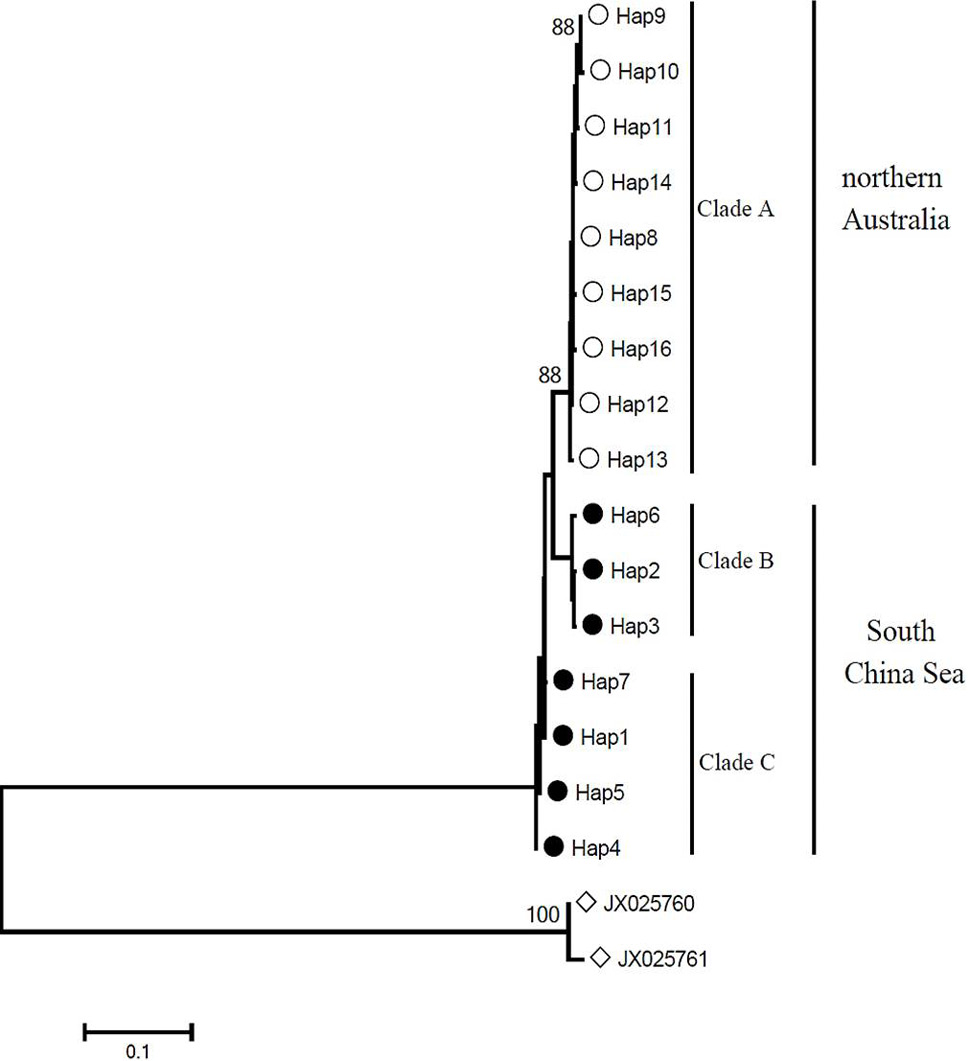
Neighbor-joining tree constructed using Tamura 3-parameter model for control region haplotypes of C. leucas. The congener C. limbatus was chosen as out-group. The black solid and hollow circles represented the South China Sea and the northern Australia population, respectively. Triangles represented the haplotype of C. limbatus. Bootstrap supports of > 85% in 1000 replicates are shown.
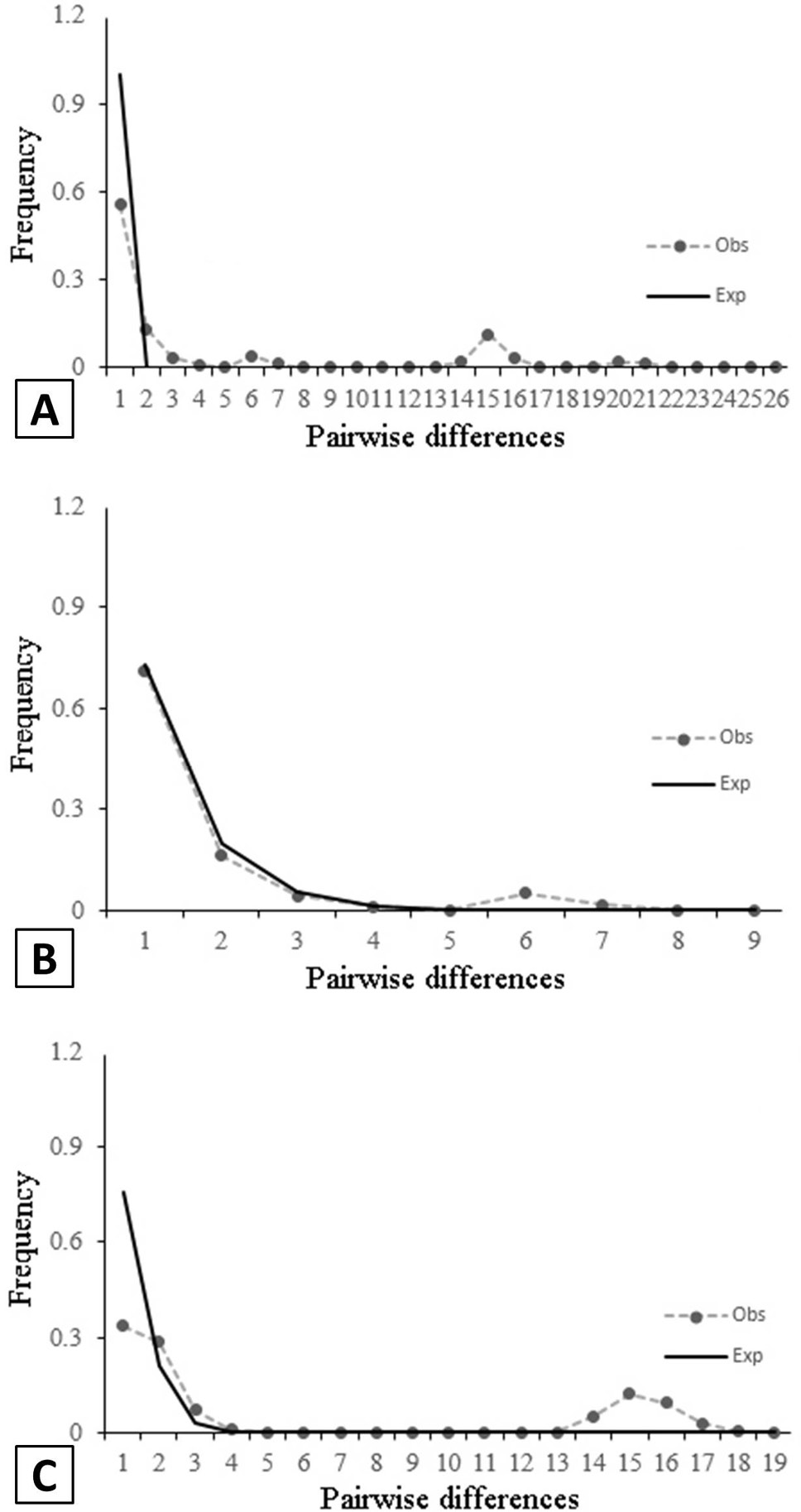
Mismatch distribution of all populations (A), northern Australia populations (B) and South China Sea populations (C). The solid line represents the curve expected (Exp.) based on the expansion model. Obs., observed.