Full-Length Genome of an Africa-4 Lineage Wild-Type Lyssavirus rabies from a Stray Dog in Egypt, 2019
Full-Length Genome of an Africa-4 Lineage Wild-Type Lyssavirus rabies from a Stray Dog in Egypt, 2019
Amthal Ahmed Fouad1, Basem Mohamed Ahmed2, Momtaz Abdelhady Shahein1, Hussein Aly Hussein2*
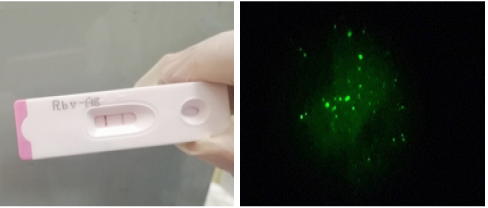
A) positive Lyssavirus rabies detection using rapid immunochromatography. B). positive Direct fluorescence indicative for RABV antigen in brain homogenate.
G-L intergenic space nucleotide features. The red box identifies the variable region where adenine (A) residues are shown to be inserted progressively. Black boxes identify the G-L TTP (G mRNA termination polyadenylation signal (left) and L mRNA transcription initiation signal (right)).
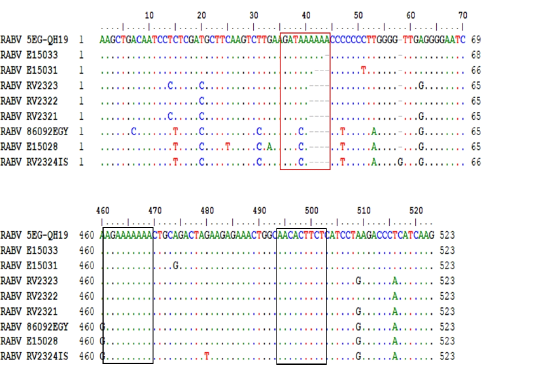
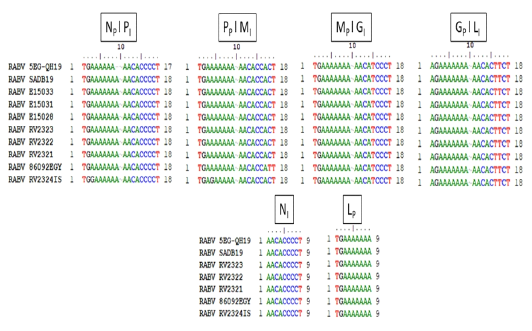
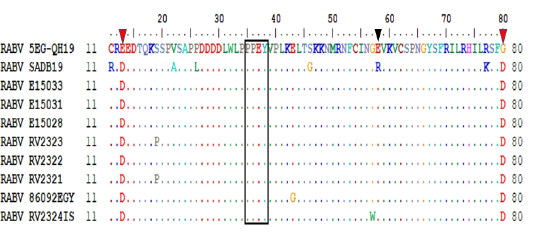
Transcription initiation and termination polyadenylation signals of 5EG-QH19 compared to SAD B19 and other genomes from AF4 lineage. Genes are presented as single letters (N-P-M-G-L). Subscript I stand for initiation while subscript P stand for polyadenylation.
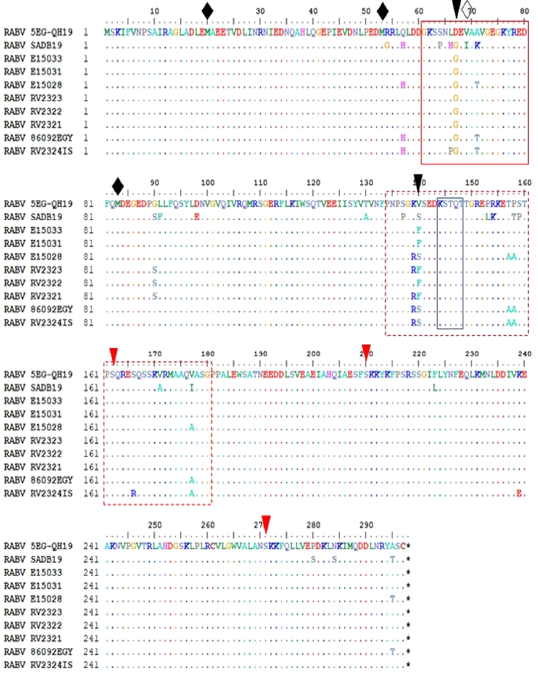
amino acid features of N. red box identify RNA binding domain. Black dashed box identifies antigenic site I. black solid boxes identify antigenic site IV. Black arrowhead for the 389S phosphorylation residue.
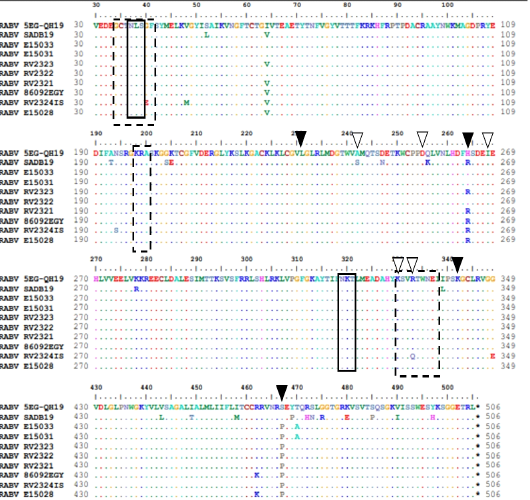
amino acid features of P protein. Variable domains (VD1) marked by red box, (VD2) marked by dashed red box. Black arrow heads indicate unique substitution in the studied sequence. Red arrowheads indicate P phosphorylation residues. Solid diamonds indicate in-frame M residues while empty diamond indicate the disrupted M69V.
5EG-QH19 M protein N-terminus showing the proline rich budding motif (black box), E58R substitution (black arrowhead) and the two unique mutations (red arrowheads).
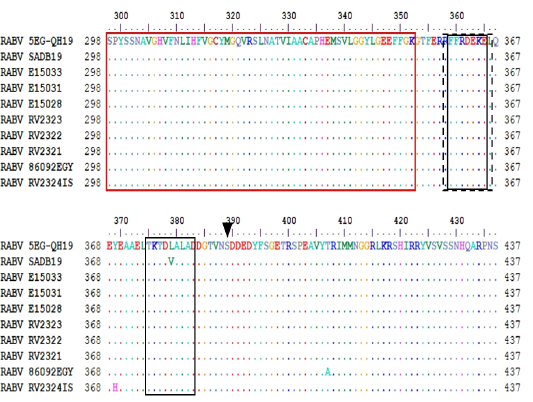
Structural features of G protein. Solid boxes show glycosylation sites. Dashed boxes for antigenic sites II and III. Black arrow heads indicate remaining antigenic sites.White arrowheads indicate pathogenicity related residues.
functional domains and carboxyl end of the L protein, RNA binding domain (543-563), polymerase active site (726-731), and polyadenylation/protein kinase related glycine-rich domain (1704-1708) in boxes, respectively.
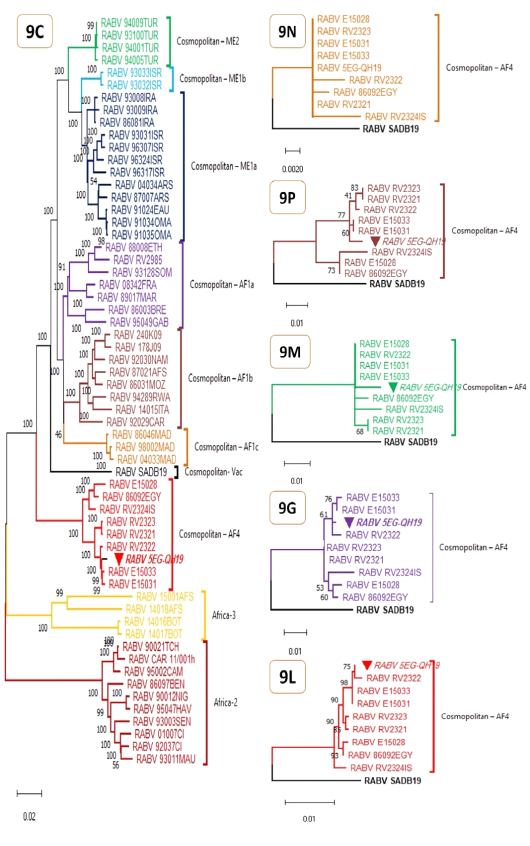
Maximum Likelihood phylogenetic analyses for complete RABV genomes (9C) and protein genes (9N, 9P, 9M, 9G, 9L). The evolutionary history was inferred by using Tamura-Nei model (Tamura and Nei 1993) for genomic tree (9C) or JTT matrix-based model (Jones et al. 1992) for protein genes (9N, 9P, 9M, 9G, 9L). The percentage of trees in which the associated taxa clustered together is shown next to the branches and is supported by 1K bootstrap test replicates for 9C and 100 bootstrap test replicates for 9N, 9P, 9M, 9G, and 9L. Initial tree(s) were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using either the Tamura-Nei model or the JTT model, and then the topology with superior log likelihood value is selected. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. All trees are midpoint rooted and clade/ lineage name is shown right to clusters. These analyses involved 60 nucleotide sequences (table S1) and 11740 positions (9C), or 10 amino acid sequences and 450, 279, 202, 524, and 2127 positions for 9N, 9P, 9M, 9G, and 9L respectively. The studied genome and its genes are marked with inverted solid triangle. Evolutionary analyses were conducted in MEGA11 (Tamura et al. 2021).