First Molecular Evidence of Anaplasma marginale Infection in Naturally Infected Cattle in Myanmar with Severe Hemolytic Anemia
First Molecular Evidence of Anaplasma marginale Infection in Naturally Infected Cattle in Myanmar with Severe Hemolytic Anemia
Babi Kyi Soe1*, Toe Win Naing2, Su Lai Yee Mon1, Nay Chi Nway3, Hiroshi Sato4
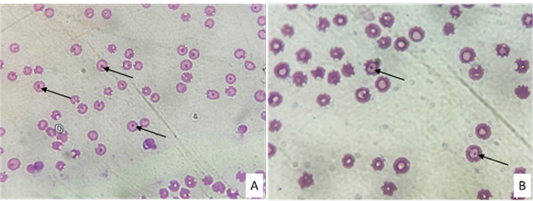
A and B showed Anaplasma inclusions (arrowhead) in the erythrocytes of Giemsa-stained thin blood smear.
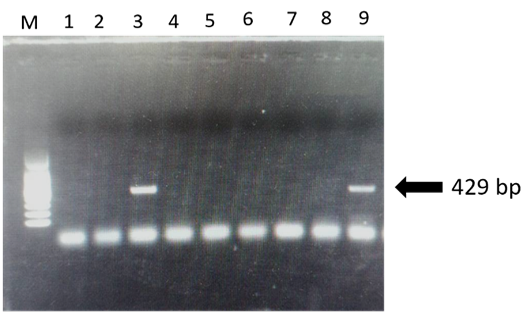
Gel electrophoresis results for PCR of Anaplasma spp. (a): M = 100bp marker, lane 3 and 9 = positive samples, and lanes 1, 2, 4, 5, 6, 7 and 8 are negative samples.
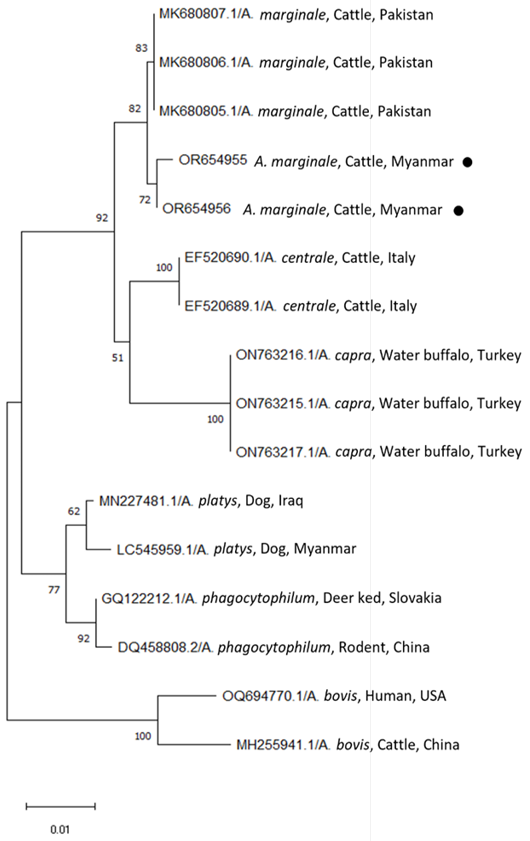
Phylogenetic trees based on the 16S rRNA sequences of Anaplasma spp. were compared with the neighbor-joining method using the MEGA software (version 11) with maximum likelihood analysis based on the general time-reversible model. Phylogenetic tree showing taxonomic relationships of resulted two sequences; OR654955 and OR654956 (solid red circle) detected from cattle in Yangon region and 14 reference sequences of 16S rRNA of all Anaplasma spp. which can be infected to cattle. The relationships were inferred based on 16S rRNA data, including 1 from dog in Myanmar and 13 reference sequences of other Anaplasma spp. originated from different countries. For each sequence, the accession numbers are followed by full species names, the host and the country of origin.