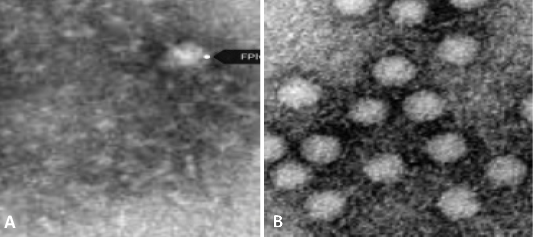
Feline Pan Leukopenia Molecular Detection and Viral Phylogeny in Egypt
Feline Pan Leukopenia Molecular Detection and Viral Phylogeny in Egypt
Omnia M. Khattab1, Morcos I. Yanni2, Hala K. Abdelmegeed2, Mahmoud Eliwa1, Naglaa M. Hagag1, Sara M. Elnomrosy1
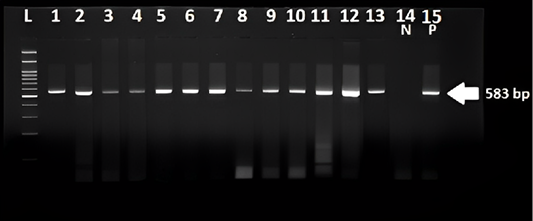
specific band with expected size 583 bp were observed. Lanes L ladder 100 pb, lane 1 to 13 were positive Samples Lane 14 negative control. Lane15 is a positive control.
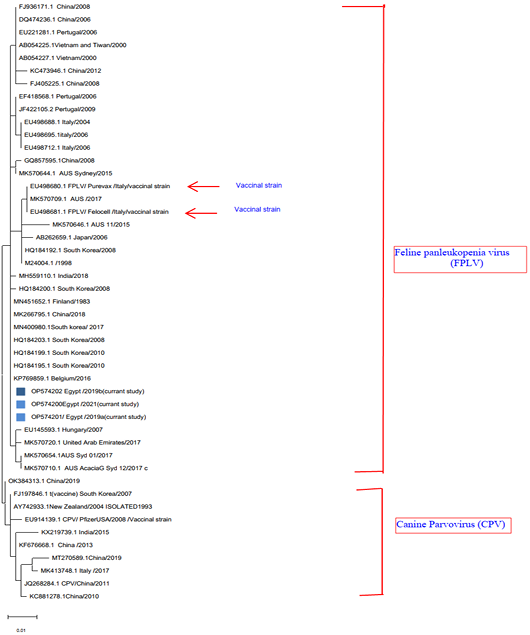
A phylogenetic tree of Feline panleukopenia based on partial VP2 gene conducted in MEGA11 (Tamura et al., 2021) using Maximum Likelihood method and Tamura-Nei model (Tamura and Nei, 1993). The tree with the highest log likelihood (−1930.82) is shown. The colour blue represents the FLP strain isolated in this study. Gen Bank IDs are located in each sequence, vaccine strains were indicated by arrow. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorthma to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 47 amino acid sequences. There was a total of 526 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021).
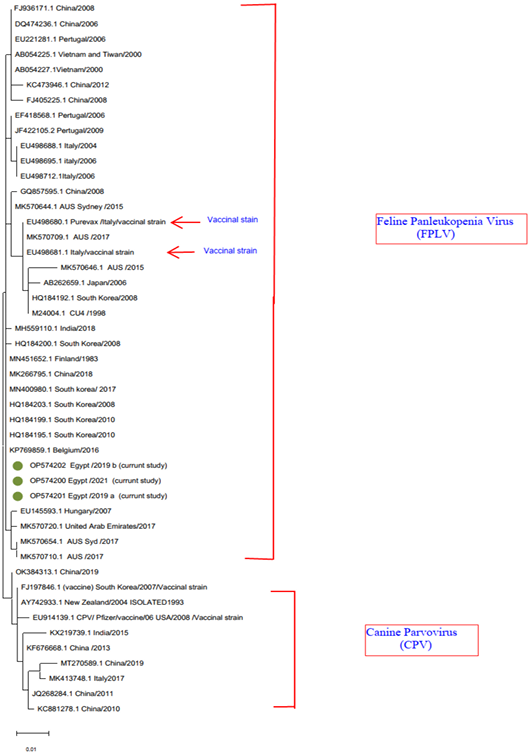
A phylogenetic tree was created using the partial sequence of VP2’s deduced amino acid sequence. The vaccine strain is shown by arrow on the tree, while the FPLV strains used in this study are shown in green. The accession numbers with brief GenBank ID are located for each sequence.
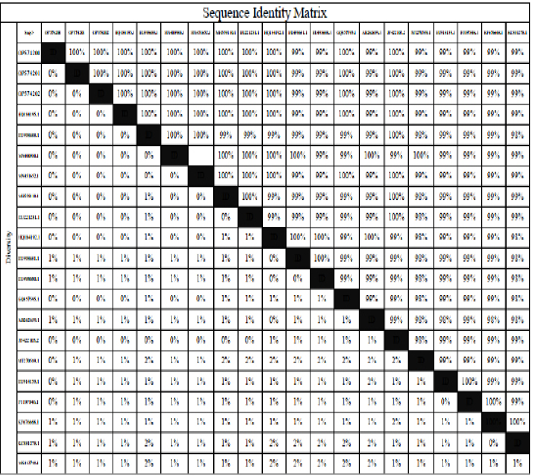
Identity percent of the recently isolated strains gene bank accession numbers OP574200, OP574201, and OP574202 and isolates illustrated in table 4 and vaccine strain used in Egypt.