Fatty Acyl-CoA Reductase of Ericerus pela (Hemiptera: Coccoidae): Localization in Insect Cells and Bioinformatic Analysis
Fatty Acyl-CoA Reductase of Ericerus pela (Hemiptera: Coccoidae): Localization in Insect Cells and Bioinformatic Analysis
Yanhong Hu* and Linkai Cui
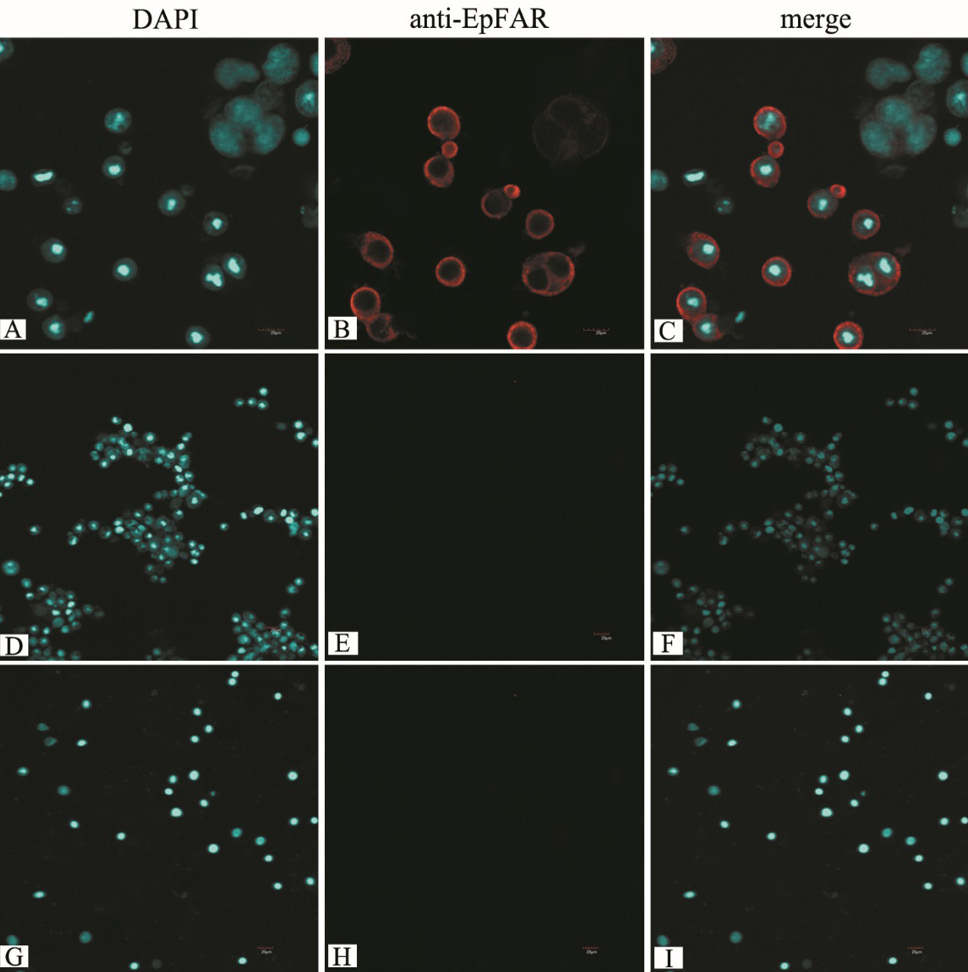
Localization of EpFAR in Sf9 cells. Sf9 cells were infected with the recombinant baculovirus and fluorescence imaged by LSCM. A, B, and C, cells were infected with the recombinant nuclear polyhedrosis baculovirus including EpFAR. D, E, and F, cells treated with the virus generated with an empty pFastBac HT B vector were used as a control. G, H, and I, cells cultured in the normal culture medium were used as another control.
FAR amino acid sequence alignment. Alignment of EpFAR and other known amino acid sequences corresponding to the sequences specified in the Table I. The ER membrane retention signals are marked with a box. On top of the alignment, the regions corresponding to the predicted transmembrane domains (TMH) of EpFAR are marked as follows: TMpred TMH are marked with a solid line (top: 23-39aa, 256-276aa, 354-372aa, 377-395aa, 500-520aa), predictions by TOPCONS are marked with a dotted lines (second from the top: 22-42aa, 379-399aa, and 498-518aa), while the TMHMM predictions are marked with dashed line (bottom: 494-516aa).