Establishment of LAMP Assay for Detection of Bacillus cereus
Establishment of LAMP Assay for Detection of Bacillus cereus
Xu Yun-Ming1*, Sun Zhi-Yuan1, Yang Jian-Bo1, Bian Rong-Rong2, Ren Hong-Lin3, Zhong Si-Yuan1, Cai Yu-Hong1, Peng Jing1 and Bao Hua-Xia1
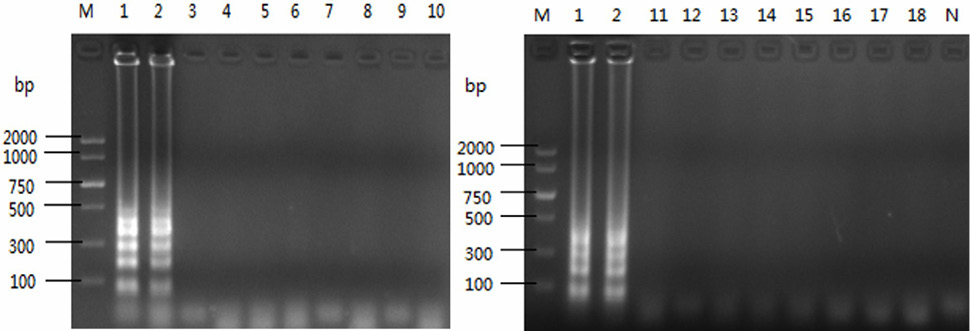
Detection specificity of LAMP reaction for strains. M, DL2000 DNA marker; N, negative control. 1-2, Bacillus cereus; 3, Yersinia enterocolitica; 4, Vibrio prholyticus; 5, Candida albicans; 6, Salmonella choleraesuls; 7, Corynebacterium glutamicum; 8, Staphylococcus aureus; 9, Micrococcus lysozyme; 10, Bacillus subtilis; 11, Escherichia coli; 12, Micrococcus luteus; 13, Edwardsiella tarda; 14, Salmonella paratyphosa; 15, Salmonella typhi; 16, Salmonella Schott; 17, Salmonella gallinarum; 18, Salmonella paratyphosa.
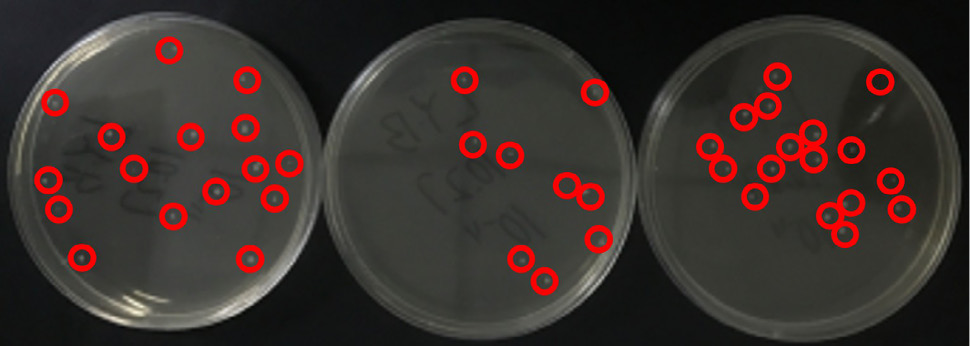
the colony counts of 10-8 times diluted B. cereus. The average colony count of the three plates was 14 CFU/mL.
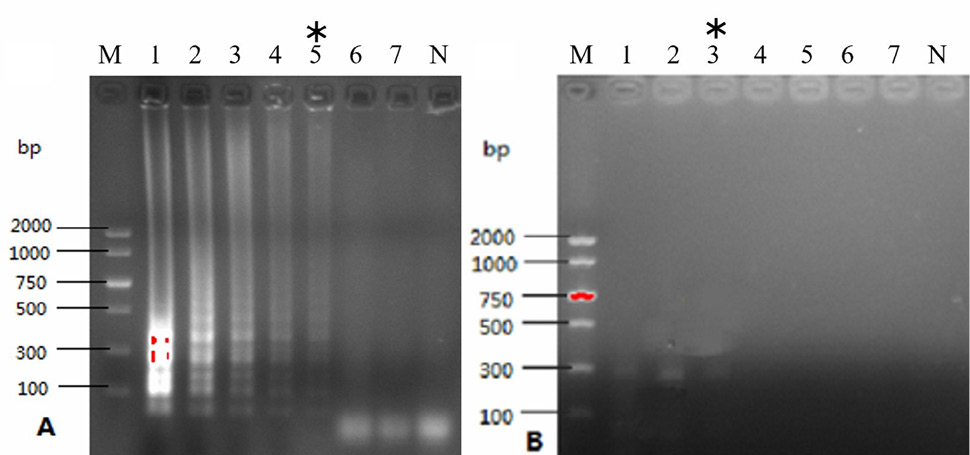
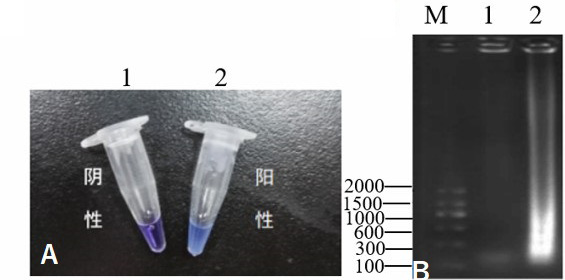
The results of HNB used in LAMP assay. M, DL2000 DNA marker; 1, negative control; 2, HNB used in LAMP assay. A, indicated results of HNB in LAMP assay with naked eye; B, electrophoresis results of HNB in LAMP assay.