Effects of Genetic and Environmental Factors on Mastitis and Performance Traits in Various Breeds of Dairy Cattle Maintained at Khyber Pakhtunkhwa, Pakistan
Effects of Genetic and Environmental Factors on Mastitis and Performance Traits in Various Breeds of Dairy Cattle Maintained at Khyber Pakhtunkhwa, Pakistan
R. Javed1,2, T. Usman1, S. Niaz2, N. Ali2,3, H. Baneh4, K. Ullah5, I. Khattak1, M. Kamal1,2,
S. Bahadar6, N.U. Khan1, S. Khan1,7, Y. Wang3*, Y. Yu3* and Mostafa K. Nassar8
1College of Veterinary Sciences and Animal Husbandry, Abdul Wali Khan University, Mardan, Pakistan
2Department of Zoology, Abdul Wali Khan University, Mardan, Pakistan
3Key Laboratory of Animal Genetics, College of Animal Science and Technology, China Agriculture University, Beijing, China
4Project Center for Agro Technologies, Skolkovo Institute of Science and Technology (Skoltech), Moscow, Russia.
5Department of Zoology, Kohat University of Science and Technology, Kohat, Pakistan
6Department of Animal Health, The University of Agriculture, Peshawar, Pakistan
7Department of Biotechnology, Abdul Wali Khan University, Mardan, Pakistan
8Department of Animal Production, Faculty of Agriculture, Cairo University, Giza, Egypt
R. Javed and N. Ali contributed equally to this study.
ABSTRACT
Milk quantity and quality traits in cattle are affected by udder health, breed, genetic (such as heritability, repeatability, and heterosis), and environmental factors. The present study was designed to evaluate the effect of udder health, genetic factors and environmental factors on milk performance traits in cattle breeds. The study was conducted on four dairy farms in Khyber Pakhtunkhwa, Pakistan. A total of 291 milk samples and 547 records were collected from lactating cattle ranging between parities one to six. For milk performance traits and udder health, each milk sample was analysed for milk composition (fat %, protein %, and lactose %) and SSC (by direct microscopic SCC method). The data were analysed using a generalized linear model of SAS studio for the effect of the SCC, breed, and herd on milk performance while wombat and CFC for the analysis of genetic and non-genetic factors. The results showed that animals with higher SCC have significantly lower (P<0.01) milk fat %. Breed-wise analysis showed significantly higher (P<0.01) protein % in Achai, lactose % in Jersey and its crossbred (Jersey x Achai), and LMY was recorded significantly higher in Friesian and its crossbred Jersey x Friesian (JF). The analysis of the environmental effect on milk composition and LMY showed that animals kept in the different herds have significantly different (P<0.01) milk compositions and LMY. The estimates of additive genetic variance (Va), variance of the permanent environment (Vpe), and estimates of phenotypic variation (Vp) were recorded highest for SCC. The analysis of the effect of genetic factors (i.e. heritability, repeatability, and heterosis) revealed that estimation of heritability was highest (0.59) for age at first calving (AFC) while lowest (0.00) for calving interval (CI), and estimation of repeatability was highest (0.99) for AFC while lowest (0.00) for CI. The results of heterosis analysis showed that crossbred (Achai x Jersey) had positive heterosis for milk composition traits and SCC while negative heterosis for LMY compared to their purebreds. The study concluded that crossbred, better herd management and nutrition, and selection based on genetic factors (such as heritability, repeatability, and heterosis) can bring improvement in milk performance traits and udder health.
Article Information
Received 07 December 2021
Revised 15 August 2022
Accepted 20 September 2022
Available online 27 April 2023
(early access)
Published 14 June 2024
Authors’ Contribution
Conceptualisation: TU, YY, YW, SN. Methodology: RJ, NA, MK, HB. Software: NA, HB. Formal analysis: RJ, IK. Visualisation: NUK, SK, MKN. Writing original draft: SB. Writing review and editing: KU. All authors have read and agreed to the published version of the manuscript.
Key words
Dairy cattle, Heritability, Mastitis, Repeatability, Somatic cell count
DOI: https://dx.doi.org/10.17582/journal.pjz/20211207121211
* Corresponding author: tahirusman@awkum.edu.pk, wangyachun@cau.edu.cn, yuying@cau.edu.cn
0030-9923/2024/0004-1909 $ 9.00/0
Copyright 2024 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Pakistan is rich in livestock and is ranked 4th in terms of milk production worldwide (Rahman et al., 2019). The annual milk production in Pakistan is about 44,977 thousand tonnes (Aujla and Hussain, 2015). Though the annual milk production is high, however, the milk production per head is quite low compared to the developed countries (Usman et al., 2014). Many factors cause low productivity per head, including late age at maturity, lower genetic potential of the dairy cattle, nutrition status, longer calving intervals, inadequate artificial insemination, and various diseases i.e. mastitis (Usman et al. 2012).
Mastitis is an inflammatory disease of lactating animals that affects udder health resulting in a reduction in the quality and quantity of milk (Usman et al., 2016). This disease manifests in two forms, i.e. clinical (CM) and sub-clinical mastitis (SCM). Mastitis causes huge economic losses to dairy farmers and the dairy industry worldwide. The higher rate of clinical mastitis is also linked with lower reproductive performance (Ali et al., 2020). Somatic cells are normal constituents of milk, but a high level of somatic cell count (SCC) in milk affects its quality, quantity and price. SCC or log-transformed somatic cell score (SCS) are useful indicators of mastitis and could be used in selection programs to lower the incidence of mastitis (Pant et al., 2007; Usman et al., 2015). To combat the per capita lower milk production in indigenous non-descript dairy cattle, the government of Pakistan imported high yielding cattle i.e. Holstein Friesian and Jersey from the western world. The crossbreeding of local breed with Holstein-Friesian shows a much faster rate of improvement in the milk production (Usman et al., 2012). Though, milk production improved by crossing these high yielding exotic breeds with local cattle, however, mastitis was also seen with the increasing trend in these exotic and crossbred cattle (Khan et al., 2019).
The estimates of genetic parameters (i.e. heritability and repeatability) of different production and reproduction traits, and genetic correlations among them are necessary to formulate the effective breeding procedures and for the estimation of breeding values (Jadoa et al., 2011). For an individual trait, the genetic variability is calculated by heritability estimates of a trait in certain environmental conditions in a herd or population (Goshu et al., 2014). Repeatability is a ratio of an individual’s inferiority or superiority for a specific trait, which is expected to be shown in future life, i.e. dry period, lactation milk yield (LMY), calving interval (CI), and lactation length, etc. It is an essential parameter to estimate the lifetime performance of an individual (Cilek and Sahin, 2009).
Seasonal variation and harsh environmental conditions in tropical and subtropical regions badly affect the performance traits of dairy cattle that results in great economic loss to the dairy industry worldwide (Linington, 1990). The true genetic ability of an animal may be suppressed by environmental factors that lead to favouritism in the selection of animals. The outcome of genetics and environment interaction (G×E) may decrease a selection response or render selection programs economically less effective (Montaldo, 2001). Heterosis is an important parameter, used in the breeding programs for better selection of desirable traits and has shown improvements in productive, reproductive, and growth traits (Lembeye et al., 2016).
In order to improve the milk production traits under subtropical conditions, it is, therefore, necessary to evaluate the effects of both genetic and non-genetic factor on incidence of mastitis and milk yield traits in dairy cattle. The present study aimed to evaluate the effect of different genetic (i.e. estimates of heritability and repeatability, and heterosis) and non-genetic factors (i.e. breed, herd, season and year) on the mastitis related traits and milk performance traits in dairy cattle maintained at various dairy farms in Khyber Pakhtunkhwa, Pakistan.
Materials and Methods
Study area and study population
The current study was conducted on four dairy farms in district Peshawar and Charsadda, Khyber Pakhtunkhwa (KP), Pakistan, from 2017 to 2019. The data for the study were obtained from dairy farms (The University of Agriculture Peshawar Dairy Farm, Government Cattle Breeding and Dairy Farm Harichand, Livestock Research and Development Station Surezai, and a private dairy farm at Khazana Charsadda. The animals belonged to three true breeds i.e. exotic Jersey (J), exotic Friesian (F), and an indigenous dairy breed Achai (A), and two crossbred cattle (i.e. FxJ and JxA). The experiments were approved by the research ethical committee of the College of Veterinary Sciences and Animal Husbandry, Abdul Wali Khan University, Mardan.
Collection of data
The dairy farms were carefully chosen on the basis of having a reliable data recording system of reproduction, production, diagnosis, and treatment of mastitis in last three years. A total of 547 complete records of 291 lactating cows was obtained from these farms, ranging between parities one to six. The primary data were collected from livestock register and then arranged in Microsoft Excel (v. 2016). The data file included the date of birth, pedigree information, date of service, date of calving, age at puberty, age at first calving, calving interval, lactation milk yield, and lactation length.
Milk sampling and analysis
Milk samples (30 mL) were collected in a sterilized bottle from all four teats of each cattle. The milk samples were carried in ice box to the Animal Breeding and Genetics Laboratory of the College of Veterinary Sciences and Animal Husbandry, Abdul Wali Khan University Mardan for analysis of milk composition, and somatic cell count (SCC).
Analysis of milk composition and SCC
Each milk sample was analysed for composition, such as fat (%), protein (%), lactose (%), and solid non-fat (SNF) using a milk analyser (Lactoscan SА standard 1060). SCC was done using direct microscopic somatic cell count method (DMSCC) as described by Ali et al. (2020).
Statistical analysis
For analysis of the effect of different factors on milk performance traits the data were analysed using generalized linear models (GLM) in SAS studio (V 9.4) for the analysis of the effects of SCC, breed, and herd on milk performance traits.
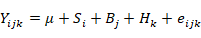
Where Yij is represents fat, protein, lactose, LMY, and SCC, µ is the overall population mean, Si is is fixed effect of ith somatic cell count (i = SCC), B j is is fixed effect of jth breed (j=1, 2, 3, 4, 5, 6), Hk is fixed effect of kth herd (k=1, 2, 3, 4) and eijK is the random error effect of each observation.
For analysis of the effects of productive and reproductive traits different parameters that effect productive and reproductive traits were analysed using the following models in Wombat. The model for calving interval (CI), fat (F %), somatic cell count (SCC), somatic cell score (SCS) and service (Ser) was:
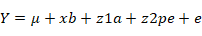
The model for age at first calving (AFC) was:
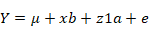
Where Y is a vector of observations, µ is average mean, xb is a vector of fixed effects such as Season (S), parity (P), herd (H) with an incidence matrix X, z1a is additive variance with incidence matrix Z, z2pe is a vector of random permanent environmental effects and e is error
To study the effect of season of calving on different traits, the year was divided into four seasons as described by Zeb et al. (2020).
Heritability (h2) were calculated using the following model.
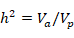
Where h2 is heritability, V a is additive genetic variance, Vp is phenotypic variation.
For estimation of repeatability (r) following statistical model was used:
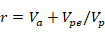
Where r is repeatability, Va is additive genetic variance, Vpe is a vector of random permanent environmental effect, Vp is phenotypic variation.
Heterosis was calculated by the following formula (Spangler, 2007).
Heterosis % = [(crossbred average – purebred average) / purebred average] x 100
Results
Association of somatic cell count with milk composition and lactation milk yield
The effect of SCC groups on milk composition traits and lactation milk yield (LMY) was observed in different cattle breeds. The results showed that animals with higher SCC (group 3) had a significantly (P<0.01) lower fat (Table I).
Association of breed with milk composition and lactation milk yield
Breed-wise analysis showed significant (P<0.01) variation in protein, lactose and LMY. Protein was found significantly higher in Achai compared to Friesian, lactose was significantly higher in Jersey and Jersey x Achai than Friesian, and LMY was recorded significantly highest in Friesian and crossbred Jersey x Friesian (Table II).
Association of herd with milk composition and lactation milk yield (LMY)
The comparison of four herds included in our study revealed that herd significantly affects milk composition
Table I. Association of SCC with milk composition and LMY.
|
SCC groups |
Fat (%) |
Protein (%) |
Lactose (%) |
SNF (%) |
LMY (Litre) |
|
1 |
4.8±0.61 A |
3.28±0.1 |
4.88±0.16 |
8.84±0.32 |
1727.51±327.75 |
|
2 |
4.89±0.67 A |
3.19±0.11 |
4.72±0.17 |
8.58±0.35 |
1730.36±341.13 |
|
3 |
3.31±0.72 B |
3.29±0.12 |
4.85±0.18 |
8.93±0.38 |
2065.04±383.12 |
|
P value |
<0.01 |
0.32 |
0.24 |
0.29 |
0.24 |
SCC groups: (Group 1, SCC <200,000 cells/mL; group 2, SCC between 200,000-500,000 cells/mL; and group 3 SCC >500,000 cells/mL. SNF, solid non-fats; LMY, Lactation milk yield. Bold P values shows a significant difference (P<0.01). Values in column with different superscript are significantly different (P<0.01).
Table II. Breed-wise comparison of milk composition and lactation milk yield (LMY).
|
Breed |
Fat (%) |
Protein (%) |
Lactose (%) |
SNF (%) |
LMY (Liter) |
|
A |
4.41±0.71 |
3.3±0.12 A |
4.9±0.18 A,B |
8.58±0.37 |
|
|
F |
4.02±0.61 |
2.97±0.1 B |
4.43±0.16 A |
8.26±0.32 |
|
|
J |
5.02±0.64 |
3.19±0.11 A |
4.81±0.16 B |
8.77±0.34 |
|
|
JA |
5.35±0.77 |
3.29±0.13 A |
4.9±0.2 B |
8.97±0.41 |
|
|
JF |
1.94±1.35 |
3.49±0.23 A,B |
5.1±0.35 A,B |
9.13±0.72 |
|
|
P value |
0.15 |
<0.01 |
<0.01 |
0.20 |
<0.01 |
Breed A, Achai; F, Friesian; J, Jersey; JA, Jersey x Achai; JF, Jersey x Friesian. For abbreviations and statistical detail, see Table I.
Table III. Herd-wise comparison of milk composition and lactation milk yield (LMY).
|
Herd |
Fat (%) |
Protein (%) |
Lactose (%) |
SNF (%) |
LMY (Liter) |
|
1 |
5±0.69 A |
3.19±0.12 A |
4.50±0.18 A |
8.42±0.37 A |
2255.21±1407.13 A |
|
2 |
3.7±0.63 B |
3.23±0.11 A |
4.62±0.16 A |
8.49±0.34 A |
1553.8±1321.6 B |
|
3 |
3.26±0.89 A,B |
3.03±0.15 A |
4.46±0.23 A |
8.57±0.47 A |
567.167±280.55 C |
|
4 |
4.68±0.78 A,B |
3.69±0.13 B |
5.50±0.20 B |
9.87±0.41 B |
3138.467±933.79 D |
|
P value |
0.01 |
<0.01 |
<0.01 |
<0.01 |
<0.01 |
Herd 1, Agriculture; 2, Harichand; 3, Surezai; 4, Khazana. For abbreviations and statistical detail, see Table I.
traits and LMY (P<0.01). Fat (%) was highest in herd 1 (Agriculture Farm, Peshawar) while protein (%), lactose (%), SNF, and LMY were highest in herd 4 (Harichand Dairy Farm, Charsadda) (Table III).
Table IV. Estimation of variance components, heritability (h2) and repeatability (r) for traits.
|
Traits |
Va |
Vpe |
Vp |
h2 |
r |
|
SCC |
13088.2 |
244834 |
498611 |
0.03 |
0.52 |
|
SCS |
0.66 |
1.39 |
2.46 |
0.27 |
0.83 |
|
AFC |
8043 |
11442 |
19485.5 |
0.59 |
0.99 |
|
Ser |
0.83 |
0.21 |
1.48 |
0.06 |
0.69 |
|
CI |
0.25 |
0.15 |
30323.9 |
0.000 |
0.00 |
|
F % |
0.32 |
0.59 |
0.95 |
0.34 |
0.96 |
SCC, somatic cell count; SCS, somatic cell score; AFC, age at first calving; Ser, service; CI, calving interval; F %, fat percentage; Va, additive variance; Vpe, variance of permanent environment; Vp, phenotypic variation; h2, heritability; r, repeatability.
Estimation of different variance, heritability (h2), and repeatability (r)
Estimates of additive genetic variance (Va) for SCC were highest (13088) while lowest for calving interval (CI). Estimates of variance of permanent environment (Vpe) were highest (244834) for SCC while lowest (0.59) for fat %. Estimates of phenotypic variation (Vp) were highest (498611) for SCC while lowest (0.95) for fat %. Estimation of heritability (h2) was highest (0.59) for age at first calving (AFC) while lowest (0.0) for CI, and estimation of repeatability (r) was highest (0.99) for AFC while lowest (0.0) for CI (Table IV).
Estimation of heterosis
Results of heterosis analysis for Achai x Jersey breed revealed positive heterosis for milk composition traits (fat %, protein %, lactose %, and SNF) and udder health (SCC) but a negative heterosis for LMY (-10.04 %) compared to its pure breeds (Achai and Jersey) (Table V). The other crossbreds (i.e. JxF and SxF) were not included in the estimation of heterosis due to their low number of animals.
Table V. Individual heterosis for Achai x Jersey.
|
Traits |
Mean for pure bred cattle (Achai and Jersey) (n=80) |
Mean for AxJ (n=18) |
Observed difference |
Heterosis (%) |
|
F % |
3.69 |
4.41 |
+0.72 |
19.67 |
|
SNF |
8.23 |
8.7 |
+0.47 |
5.78 |
|
P % |
3.12 |
3.22 |
+0.1 |
3.37 |
|
L % |
4.67 |
4.85 |
+0.18 |
3.97 |
|
LMY (l) |
804.72 |
723.94 |
-80.78 |
-10.04 |
|
SCC |
204000 |
129000 |
+75000 |
36.76 |
F %, fat percentage; SNF, solid non-fat; P %, Protein percentage; L %, lactose percentage; LMY, Lactation milk yield; SCC, somatic cell count.
Discussion
The present study evaluated the effect of genetic and environmental (non-genetic) factors on the milk performance traits and udder health in dairy cattle maintained at dairy farms in Khyber Pakhtunkhwa, Pakistan. The present study found significantly lower (P<0.01) fat (%) in cattle with higher SCC. The finding of the current study is similar to the findings of Cinar et al. (2015) and Kul et al. (2014) who recorded that high SCC negatively affects the milk yield and milk composition (fat %, SNF, protein %, and lactose %). El-Tahawy and El-Far (2010) also noted a significantly negative correlation between SCC and milk performance. The current study recorded increased protein % in cows with higher SCC, though the increase was non-significant. Atasever and Stádník (2015) also noted a positive correlation between protein % and SCC.
The breed-wise analysis in the current study recorded significantly higher protein (%) in the indigenous breed Achai, lactose % in Jersey and its crossbred (Jersey x Achai), and LMY was recorded significantly higher in Friesian and its crossbred (JF). Mushtaq et al. (2012) reported that breed significantly affected the fat, protein, and lactose percentages. Hassan and Khan (2013) reported similar results that total milk yield was higher for exotic crossbreds as compared to pure breed. Galukande et al. (2013) also found that crossbreeds had a higher milk yield. Uddin et al. (2014) studied production performance of Achai cattle at the Surezai dairy farm and reported a higher LMY (813.07±113.3 litre) than the present study.
In the present study farm-wise comparison showed that milk composition and LMY significantly vary in these herds. Bach et al. (2008) and Sova et al. (2013) in their studies also reported that herd management, such as herd size, feed type, feed frequency, cleaning frequency, chemical quality of drinking water, and other facilities greatly affect the milk composition, milk yield, and SCC.
The concept of heritability and repeatability coupled with genetic and environment plays an important role in variation in different performance traits. It contains the additive genetic variance, phenotypic variance, and variance of permanent environment for each animal. Low estimates of repeatability showed that traits are usually influenced by management systems and environment compared to the genotype and permanent environmental effects (Islam et al., 2017).
In the present study, estimates of additive genetic variance (Va), variance of permanent environment (Vpe), and estimates of phenotypic variation (Vp) were recorded highest for SCC, while estimates of heritability (h2) and repeatability (r) were highest for age at first calving (AFC). Guzzo et al. (2018) reported different estimates of Va, Vpe and heritability for SCC compare to the current study. Costa et al. (2019) reported a different estimate of variance components for SCS, whereas estimates for heritability and repeatability (0.10 and 0.53, respectively) recorded by their study are closer to the present study. Vinothraj et al. (2016) reported estimates for service which was different from our findings. Dash et al. (2015) reported a different estimate of heritability and repeatability for CI (0.61 and 0.10, respectively). Chegini et al. (2015) reported different heritability estimates for AFC and CI (0.13 and 0.44, respectively). The estimates of variance components (Va, Vpe, and Vp), heritability, and repeatability for fat % reported by Behzadi et al., (2013) are not in line with the current study. Studies show that higher heritability estimate increases the chances of a trait to be inherited to offsprings while better management can improve repeatability.
The heterosis results obtained in our study showed that the crossbred (AxJ) has positive heterosis for milk production traits (F %, P %, L %, and SNF %) and SCC while a negative heterosis for LMY. Kargo et al. (2021) recorded positive heterosis for protein yield and total milk yield than one of the pure breed in their study. Lembeye et al. (2016) also showed positive heterosis for production traits in crossbred (F x J) of their study.
Conclusion
The present study concluded that lower SCC and better herd management can affect the udder health that has positive impacts on milk composition and total milk yield. The exotic breed and their crossbreds have high milk production compared to the indigenous dairy breeds (Achai). The study suggests that selecting traits with higher heritability (i.e. AFC and F %) in breeding programs can bring improvements in these traits.
Acknowledgement
We are thankful to the dairy farms for providing samples and data for this study.
Declaration of Competing Interest
None.
Funding
The research was funded by the joint project of Pakistan Science Foundation and Nature Science Foundation of China PSF-NSFCIII/Agr/KP/AWKUM/(20) and (31961143009). The funders had no role in study design, data collection, analysis, decision to publish, and preparation of the manuscript.
IRB approval
The study did not involved any human subject neither it included any human or animal pathogen. Therefore, it did not require an IRB approval.
Ethical statement
The study was approved by the ethical committee of the College of Veterinary Sciences and Animal Husbandry, Abdul Wali Khan University Mardan.
Statement of conflict of interest
The authors have declared no conflict of interest.
References
Ali, N., Niaz, S., Khan, N.U., Gohar, A., Khattak, I., Dong, Y., Khattak, T., Ahmad, I., Wang, Y., and Usman, T., 2020. Polymorphisms in JAK2 gene are associated with production traits and mastitis resistance in dairy cattle. Annls Anim. Sci., 20: 409-423. https://doi.org/10.2478/aoas-2019-0082
Atasever, S., and Stádník, L., 2015. Factors affecting daily milk yield, fat and protein percentage, and somatic cell count in primiparous Holstein cows. Indian J. Anim. Res., 49: 313-316. https://doi.org/10.5958/0976-0555.2015.00048.5
Aujla, K.M., and Hussain, A., 2015. Economics of milk production of major dairy buffalo breeds by agro-ecological zones in Pakistan. Pak. J. agric. Res., 28: 179-191.
Bach, A., Valls, N., Solans, A. and Torrent, T., 2008. Associations between nondietary factors and dairy herd performance. J. Dairy Sci., 91: 3259-3267. https://doi.org/10.3168/jds.2008-1030
Behzadi, B.M.R., Amini, A., Slaminejad, A.A., and Tahmoorespour, M., 2013. Estimation of genetic parameters for production traits of Iranian Holstein dairy cattle. Livest. Res. Rural. Dev., 25.
Chegini, A., Shadparvar, A.A., and Ghavi, H.Z.N., 2015. Genetic parameter estimates for lactation curve parameters, milk yield, age at first calving, calving interval and somatic cell count in Holstein cows. Iran. J. appl. Anim. Sci., 5: 61-68.
Cilek, S., and Sahin, E., 2009. Estimation of some genetic parameters (heritability and repeatability) for milk yield in the Anatolian population of Holstein cows. Arch. Zoot., 12: 57-64.
Cinar, M., Serbester, U., Ceyhan, A., and Gorgulu, M., 2015. Effect of somatic cell count on milk yield and composition of first and second lactation dairy cows. Ital. J. Anim. Sci., 14: 3646. https://doi.org/10.4081/ijas.2015.3646
Costa, A., Lopez-Villalobos, N., Visentin, G., De Marchi, M., Cassandro, M., and Penasa, M., 2019. Heritability and repeatability of milk lactose and its relationships with traditional milk traits, somatic cell score and freezing point in Holstein cows. Animal, 13: 909-916. https://doi.org/10.1017/S1751731118002094
Dash, S.K., Gupta, A.K., Singh, A., Manoj, M., Shivahre, P.R., Panmei, A., and Sahoo, S.K., 2015. Covariance component and genetic parameter estimate of production and fertility traits in Holstein Friesian cross cattle using repeatability animal model. Indian J. Anim. Sci., 85: 1092-1097.
El-Tahawy, A.S., and El-Far, A.H., 2010. Influences of somatic cell count on milk composition and dairy farm profitability. Int. J. Dairy Technol., 63: 463-469. https://doi.org/10.1111/j.1471-0307.2010.00597.x
Galukande, E., Mulindwa, H., Wurzinger, M., Roschinsky, R., Mwai, A.O., and Sölkner, J., 2013. Crossbreeding cattle for milk production in the tropics: achievements, challenges and opportunities. Anim. Genet. Resour. Inf., 52: 111-125. https://doi.org/10.1017/S2078633612000471
Goshu, G., Singh, H., Petersson, K.J., and Lundeheim, N., 2014. Heritability and correlation among first lactation traits in Holstein Friesian cows at Holeta Bull Dam Station, Ethiopia. Int. J. Livest. Prod., 5: 47-53. https://doi.org/10.5897/IJLP2013.0165
Guzzo, N., Sartori, C., and Mantovani, R., 2018. Genetic parameters of different measures of somatic cell counts in the Rendena breed. J. Dairy Sci., 101: 8054-8062. https://doi.org/10.3168/jds.2017-14047
Hassan, F., and Khan, M.S., 2013. Performance of crossbred dairy cattle at military dairy farms in Pakistan. J. Anim. Pl. Sci., 23: 705-714.
Islam, M.S., Akhtar, A., Hossain, M.A., Rahman, M.F., and Hossain, S.S., 2017. Reproductive performance and repeatability estimation of some traits of crossbred cows in savar dairy farm. J. environ. Sci. nat. Resour., 10: 87-94. https://doi.org/10.3329/jesnr.v10i2.39017
Jadoa, A.Y.A.A.J., and Abdulrada, A.J., 2011. Heritabilties and breeding values of production and reproduction traits of Holstein cattle in Iraq. J. Basrah Res. (Sci.)., 37: 67-70.
Kargo, M., Clasen, J.B., Nielsen, H.M., Byskov, K., and Norberg, E., 2021. Heterosis and breed effects for milk production and udder health traits in crosses between Danish Holstein, Danish Red, and Danish Jersey. J. Dairy Sci., 104: 678-682. https://doi.org/10.3168/jds.2019-17866
Khan, M.Z., Wang, D., Liu, L., Usman, T., Wen, H., Zhang, R., Liu, S., Shi, L., Mi, S., Xiao, W., and Yu, Y., 2019. Significant genetic effects of JAK2 and DGAT1 mutations on milk fat content and mastitis resistance in Holsteins. J. Dairy Res., 86: 388–393. https://doi.org/10.1017/S0022029919000682
Kul, E., Şahin, A., Atasever, S., Uğurlutepe, E. and Soydaner, M., 2019. The effects of somatic cell count on milk yield and milk composition in Holstein cows. Vet. Arhiv, 89: 143-154. https://doi.org/10.24099/vet.arhiv.0168
Lembeye, F., López-Villalobos, N., Burke, J.L., and Davis, S.R., 2016. Breed and heterosis effects for milk yield traits at different production levels, lactation number and milking frequencies. N. Z. J. agric. Res., 59: 156-164. https://doi.org/10.1080/00288233.2016.1156551
Linington, M.J., 1990. The use of Sanga cattle in beef production. Tech. Commun. Dep. Agric. Dev. S. Afr., 223: 31-37.
Montaldo, H.H., 2001. Genotype by environment interactions in livestock breeding programs: A review. Interciencia, 26: 229-235.
Mushtaq, A., Qureshi, M.S., Khan, S., Habib, G., Swati, Z.A., and Rahman, S.U., 2012. Body condition score as a marker of milk yield and composition in dairy animals. J. Anim. Pl. Sci., 22(Suppl 3): 69-173.
Pant, S.D., Schenkel, F.S., Leyva-Baca, I., Sharma, B.S., and Karrow, N.A., 2007. Identification of single nucleotide polymorphisms in bovine CARD15 and their associations with health and production traits in Canadian Holsteins. BMC Genom., 8: 1-11. https://doi.org/10.1186/1471-2164-8-421
Rahman, M.A., Saboor, A., Hameed, G., Bilal, G., and Tanwir, F., 2019. Climate change related factors impacting dairy production in Pakistan. Pak. J. agric. Sci., 32: 691-705. https://doi.org/10.17582/journal.pjar/2019/32.4.691.705
Sova, A.D., LeBlanc, S.J., McBride, B.W., and DeVries, T.J., 2013. Associations between herd-level feeding management practices, feed sorting, and milk production in freestall dairy farms. J. Dairy Sci., 96: 4759-4770. https://doi.org/10.3168/jds.2013-6679
Spangler, M.L., 2007. The value of heterosis in cow herds: Lessons from the past that apply to today. Proc. Range Beef Cow Symp. XX December 11, 12 and 13, 2007 - Fort Collins, Colorado.
Uddin, H., Khan, H.U., Khan, M.I., Khan, R., and Naveed, A., 2014. Productive and reproductive performance of Achai cattle maintained at livestock research and development station Surezai Peshawar, Pakistan. J. Anim. Hlth. Prod., 22: 2014.
Usman, T., Guo, G., Suhail, S.M., Ahmed, S., Qiaoxiang, L., Qureshi, M.S., and Wang, Y., 2012. Performance traits study of Holstein Friesian cattle under subtropical conditions. J. Anim. Pl. Sci., 22: 92-95.
Usman, T., Suhail, S.M., Zhang, X., Guo, G., Ullah, I., Sadique, U., and Wang, Y., 2014. Effects of non-genetic factors on age at first conception and calving interval in Holstein cattle under subtropical conditions. Indian J. Anim. Sci., 84: 45-49.
Usman, T., Wang, Y., Liu, C., Wang, X., Zhang, Y., and Yu, Y., 2015. Association study of single nucleotide polymorphisms in JAK 2 and STAT 5B genes and their differential mRNA expression with mastitis susceptibility in Chinese Holstein cattle. Anim. Genet., 46: 371–380. https://doi.org/10.1111/age.12306
Usman, T., Wang, Y., Song, M., Wang, X., Dong, Y., Liu, C., Wang, S., Zhang, Y., Xiao, W., and Yu, Y., 2017. Novel polymorphisms in bovine CD4 and LAG-3 genes associated with somatic cell counts of clinical mastitis cows. Genet. mol. Res., 17: gmr16039859. https://doi.org/10.4238/gmr16039859
Usman, T., Yu, Y., Zhai, L., Liu, C., Wang, X., and Wang, Y., 2016. Association of CD4 SNPs with fat percentage of Holstein cattle. Genet. mol. Res., 15: gmr.15038697. https://doi.org/10.4238/gmr.15038697
Vinothraj, S., Subramaniyan, A., Venkataramanan, R., Joseph, C, and Sivaselvam, S.N., 2016. Genetic evaluation of reproduction performance of Jersey× Red Sindhi crossbred cows. Vet. World, 9: 1012. https://doi.org/10.14202/vetworld.2016.1012-1017
Zeb, S., Ali, N., Niaz, S., Rasheed, A., Khattak, I., Khan, N.U., Wang, Y., and Usman, T., 2020. Association of SNPs in the coding regions of CD4 gene with mastitis susceptibility and production traits in dairy cattle. Thai J. Vet. Med., 50: 75-80.
To share on other social networks, click on any share button. What are these?










