Diversity Analysis of Chinese Tibetan Naqu Yak (Bos grunniens) Populations Using mtDNA
Diversity Analysis of Chinese Tibetan Naqu Yak (Bos grunniens) Populations Using mtDNA
Wang-Dui Basang1, Tian-Wu An2, Luo-Bu Danjiu3, Yan-Bin Zhu1, Shi-Cheng He3, Xiao-Lin Luo2, Wei-Wei Ni4, Xiao Wang4, Shu-Zhu Cheng4, Jian Wang4 and Guang-Xin E4,*
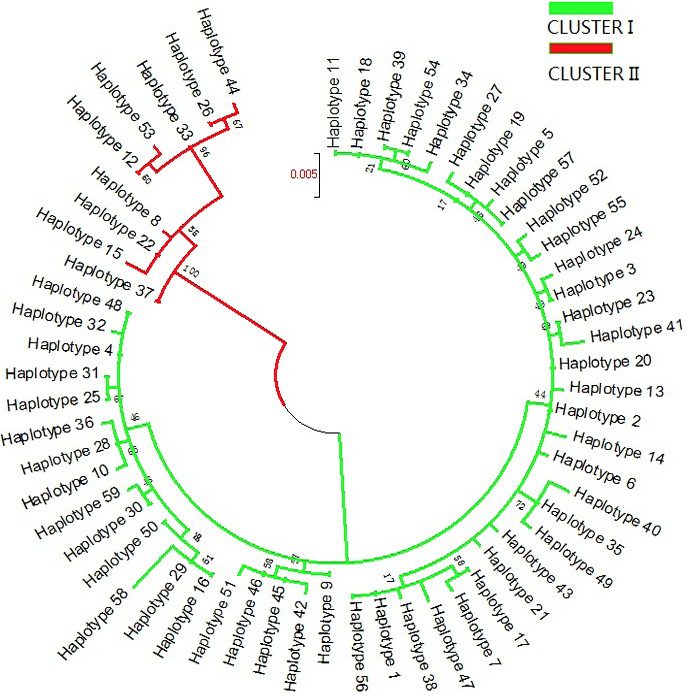
Molecular phylogenetic analysis of 59 Naqu yak mtDNA D-loop haplotypes using the maximum likelihood method. The evolutionary history was inferred using the maximum likelihood method based on the Tamura-Nei model (Tamura and Nei, 1993). The tree with the highest log likelihood (-1693.9317) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying the neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with a superior log likelihood value. The rate variation model allowed some sites to be evolutionarily invariable ([+I], 46.4858% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 59 nucleotide sequences. A total of 811 positions were included in the final dataset.
Network and frequency profiles of the 59 Naqu yak haplotypes with mtDNA D-loop sequences.
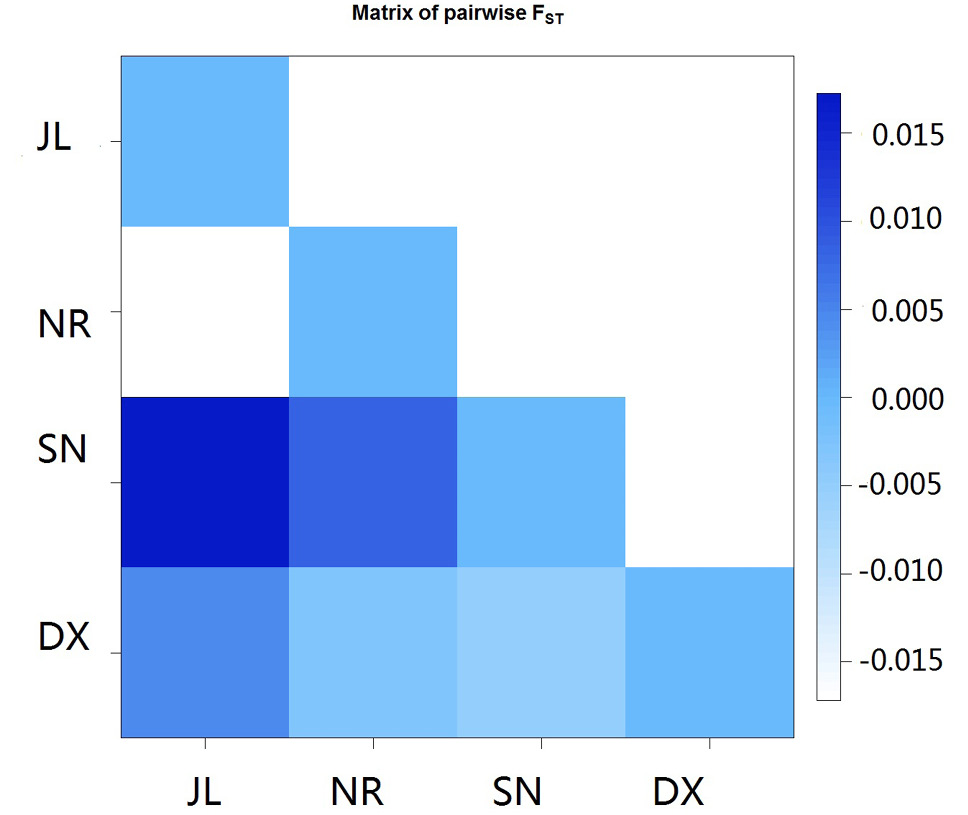
Matrix of pairwise FST values of the four Naqu ecotype yak populations.
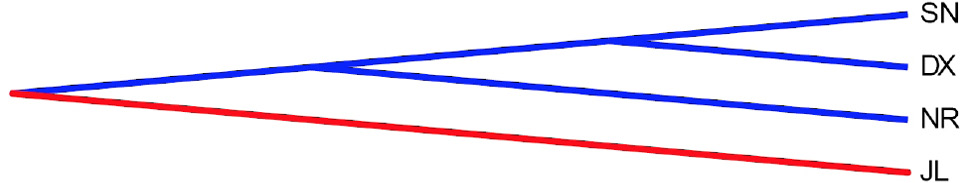
Average number of nucleotide differences between populations (Kxy) among the four Naqu ecotype Yak populations based on DnaSP 5.10.