Detection and Molecular Characterization of Virulent Newcastle Disease Virus in Ducks (Anas platyrhynchos domesticus)
Detection and Molecular Characterization of Virulent Newcastle Disease Virus in Ducks (Anas platyrhynchos domesticus)
Beenish Zahid*1,2,7, Javed Iqbal Qazi2, Amir Zohaib2, Asim Aslam3, Raheela Akhter3, Haleema Sadia4, Qurat ul Ain5, Razia Sultana6, Irfan Irshad3 and Sobia Alyas7
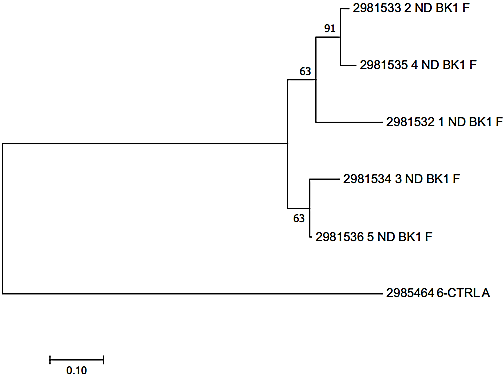
Evolutionary relationships of taxa. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 1.61196123 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The analysis involved 6 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 212 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.
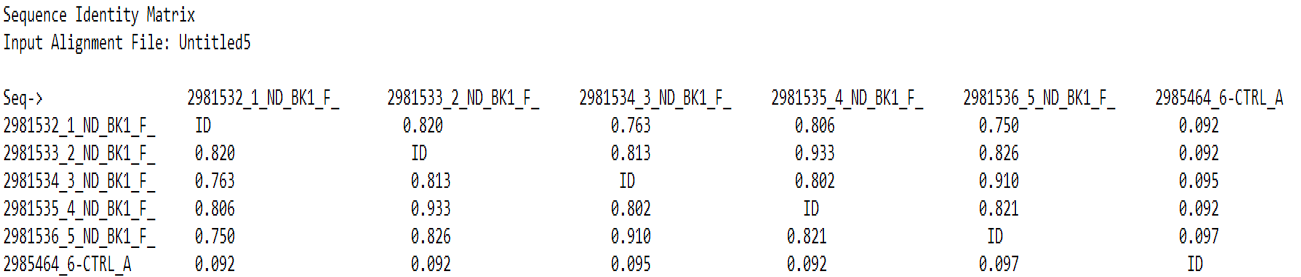
The similarity matrix of all the five NDV isolated from ducks.