Current Status of Citrus Tristeza Virus in Major Citrus Growing Areas of Sargodha, Pakistan
Research Article
Asim Ali Khan1, Muhammad Ahmad Zeshan1, Yasir Iftikhar1*, Mustansar Mubeen1, Maham Sohail1, Sonum Bashir1, Malik Abdul Rehman2 and Muhammad Usman Ghani3
1 Department of Plant Pathology, College of Agriculture, University of Sargodha, Sargodha, Pakistan; 2Citrus Research Institute, Sargodha, 40100, Pakistan; 3 Institute of Soil and Environmental Sciences, University of Agriculture Faisalabad, Pakistan.
Abstract | Tristeza disease is caused by citrus tristeza virus (CTV) expresses salient symptoms like stem pitting, yellowing of leaves, stunted growth of large, small lateral branches and twigs. The study was carried out to quantify the spatial and temporal distribution of CTV disease in major citrus growing areas of Sargodha district (Sargodha, Kot Momin, and Bhalwal). The samples from the declined orchards were collected based on symptomology and confirmed through ELISA. CTV was detected in 3 varieties viz., Kinnow (Citrus reticulate), Mosambi (Citrus sinensis) and Feutrell’s early (Citrus reticulate cv feutrell’s early). The maximum disease incidence was recorded in Mosambi (90%), followed by Feutral’s early (74%) and Kinnow (57%). Mosambi was the most susceptible within the range of OD values (0.82-2.35) followed by Feutrell’s early (0.45-0.64) and Kinnow (0.25-0.56). The disease severity was recorded only on Mosambi as it was more affected than Feutral’s early and Kinnow. The rating scale to observe the severity of tristeza disease was based on percentage of pits in the 1 sq. inch piece of bark at bud union of infected tree. This rating scale has been developed for the first time to record the tristeza disease severity. The maximum disease severity was recorded in Kot Momin (62%) followed by Bhalwal (56%) and Sargodha (50%). The study concluded that CTV is one of the most promising factors playing role in the citrus decline.
Received | February 22, 2022; Accepted | May 16, 2022; Published | October 05, 2022
*Correspondence | Yasir Iftikhar, University of Sargodha, Sargodha, Pakistan; Email: yasir.iftikhar@uos.edu.pk
Citation | Khan, A.A., M.A. Zeshan, Y. Iftikhar, M. Mubeen, M. Sohail, S. Bashir, M.A. Rehman and M.U. Ghani. 2022. Current status of citrus tristeza virus in major citrus growing areas of Sargodha, Pakistan. Sarhad Journal of Agriculture, 38(4): 1412-1418.
DOI | https://dx.doi.org/10.17582/journal.sja/2022/38.4.1412.1418
Keywords | CTV, Disease severity, citrus decline, Disease mapping, Sargodha
Copyright: 2022 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Citrus is cultivated in more than 64 countries of the world and its worldwide production is 140 million tons annually (Hussain et al., 2021; Sajid et al., 2021). Citrus tristeza virus (CTV) is an economically important pathogen because it attacks the citrus orchards throughout the world and left millions of trees unproductive or completely destroyed. Since the emergence of CTV in the 20th century, the epidemic form of CTV had destroyed more than 60 million citrus trees (Iftikhar et al., 2021). CTV belongs to the genus Closterovirus; family Closteroviridae and is transmitted very efficiently by different species of aphids (Lin et al., 2006). CTV has different strains from mild to severe and the symptoms of CTV are strain dependents (Iftikhar et al., 2021). Mild isolates of CTV cannot cause decline whereas virulent strains cause huge losses. The main symptoms are stem pitting in twigs, honeycombing, vein clearing in lime, and gummosis on small lateral branches and twigs (Iftikhar et al., 2013). CTV was first documented in Pakistan in the late 80s during a survey of citrus plantations in KPK and Punjab. It was confirmed through ELISA and electron microscopy (Lin et al., 2006). The CTV was also documented in different areas of Punjab and KPK during extensive surveys in 2006-2007 (Abbas et al., 2008; Iftikhar et al., 2009). The highest incidence of CTV (48%) was recorded in Bhalwal (Punjab) and 40% incidence was found in Mardan (KPK) (Iftikhar et al., 2009). Again in 2013, 67% incidence in Sargodha and 39% in Mardan was reported (Naseem et al., 2016). Presently, very rare studies were conducted regarding spatio-temporal dynamics of CTV disease in Sargodha which is also known as California of Pakistan. Keeping in view the above-mentioned facts about CTV, the current study was aimed to quantify the spatial and temporal distribution of CTV disease in 3 Tehsils of Sargodha because Bhalwal, Kot Momin and Sargodha are major citrus growing regions in the district. The assessment of CTV disease occurrence in 3 citrus cultivars i.e., Kinnow, Mosambi and Feutral’s early which are readily available in the mentioned areas would provide a base to map out its role in the citrus decline and to devise sustainable management options.
Materials and Methods
Survey and collection of infested samples
A survey of citrus fields was conducted in different fields of 3 major citrus growing Tehsils of Sargodha District viz., Sargodha, Bhalwal and Kot Momin where 18 orchards were surveyed from each Tehsil. This survey included 3 varieties of citrus viz., Mosambi (Citrus sinensis) Feutral’s early (Citrus reticulate cv Feutrell’s early) and Kinnow (Citrus nobilis x Citrus deliciosa). Salient features of CTV such as; dieback, stem pitting, honeycombing in bark and leaf yellowing were used as morphological markers to identify the diseased samples at initial step. The samples based on symptoms were collected in polythene bags and immediately shifted to ice bucket. The samples were collected in such a way that 6 orchards of each variety i.e. Kinnow, Mosambi and Feutrell’s early were selected in every Tehsil and 3 samples were collected from each orchard; thus 54 samples/ Tehsil were brought to the lab for further processing to confirm the virus. The orchards were selected on the basis of 3 age groups i.e., up to 5 years, 6-10 years and 11-15 years.
Extraction of virus sap
The collected samples were washed with tap water to remove the dust particles followed by 3 washings with distilled water and drying on blotter paper. The samples were crushed in a pre-chilled pestle and mortar by using extraction buffer. The extract was sieved through muslin cloth and collected in Eppendorf separately for each sample (Byamukama et al., 2013).
Confirmation of virus
The presence of Virus was confirmed through enzyme linked immunosorbent assay (ELISA). The suspected samples were subjected to double antibody sandwich (DAS) ELISA as described by Iftikhar et al. (2009) as follows.
Microtiter plates were coated with CTV-specific antibody diluted 1000-folds in coating buffer followed by incubation at 30°C for 4 hours. After the incubation, ELISA plates were washed thrice with washing buffer to remove the excessive antibodies. Freshly prepared antigens (1:10 w/v) in extraction buffer were charged (100μl/well) and the plates were again incubated overnight at 4ºC followed by repeating the washing. Reference blank, negative and positive controls were also included in the ELISA plate. CTV enzyme-conjugated antibodies were diluted 1000-fold in the conjugate and ELISA plate wells were coated by adding 100μl/well and incubated at 30°C for 5 hours followed by washing. Substrate (p-nitrophenyl-phosphate) was added @ 1mg/ml and incubated at room temperature for an hour at least. The reaction strength was rated visually as: = no reaction; + = weak reaction; ++ = definite reaction; +++ = strong reaction; ++++ = very strong reaction.
Disease incidence data recording
Disease incidence was recorded, after the confirmation of CTV through ELISA, in the samples collected from the surveyed orchards, the disease incidence data were calculated by using the following formula:
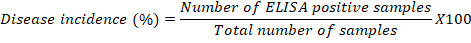
Severity of CTV
The severity of CTV was calculated in Mosambi orchards as characteristics symptoms were more pronounced in Mosambi trees affected with CTV. Based on apparent symptomology, the plants were selected from each field in a randomized pattern and 1 square inch piece of barks were taken out from bud union and the number of pits were observed. The area covered by pits was calculated out of a total 1sq inch area and determined the severity by the percentage of the pits (Table 1). The pits are salient symptoms of CTV disease while leaf yellowing and mottling etc. also resemble with other diseases, that’s why pits were selected to calculate disease severity. Among all the citrus varieties, Mosambi was the most affected citrus variety therefore, disease severity was recorded only in Mosambi.
Table 1: Percentage of pits per square inch of bark.
|
Severity level |
Pits (%) |
|
Least severe |
0-20% |
|
Slightly severe |
21-40% |
|
Moderately severe |
41-60% |
|
Severe |
61-80% |
|
Highly severe |
81-100% |
Results and Discussion
The random survey was carried out including diseased and healthy citrus orchards of Sargodha, Bhalwal and Kot Momin, where 18 orchards were surveyed from each Tehsil. Out of 18 orchards at each Tehsil 6 orchards were marked per cultivar i.e., Kinnow, Mosambi and Feutrell’s early that was further divided into three age groups where 2 diseased and 1 healthy sample was collected (Table 2).
ELISA showed the presence of the virus by developing yellow colour in the wells having positive samples (Figure 1). The optical density (OD) of the colour showed the intensity regarding the presence of virus in the wells. OD values of infected samples were compared to those of healthy and control plants (Table 2). The OD values indicated that wells with dark yellow color have more concentration of CTV as compared to light yellow colour. In Tehsil Kot Momin, the OD values for infected samples of Mosambi were ranged from 0.89 to 2.35 while healthy samples of the same area showed very low value (0.17). The OD values for infected samples of Feutrell’s early in Tehsil Kot Momin was less than Mosambi which ranged from 0.45 to 0.61 while in case of healthy samples it was 0.12. The infected samples of Kinnow showed the least OD values as compared to Mosambi and Feutrell’s early ranging from 0.28 to 0.53 which was only 0.11 in case of healthy Kinnow samples.
Table 2: The survey of citrus orchards.
|
Cultivar |
Tehsils |
||||||||
|
Bhalwal |
Kot Momin |
Sargodha |
|||||||
|
Kinnow |
Mosambi |
FE* |
Kinnow |
Mosambi |
FE |
Kinnow |
Mosambi |
FE |
|
|
AG 1** |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
|
AG 2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
|
AG 3 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
|
Total |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
|
S/O*** |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
|
T/C^ |
18 |
18 |
18 |
18 |
18 |
18 |
18 |
18 |
18 |
|
GT |
18 |
18 |
18 |
||||||
*FE= Feutral’s early; **AG= Age group; AG1= up to 5 years; AG2= 6-10 years; AG3= 11-15 years; ***S/O = Number of samples per orchard; ^T/C = Number of samples per cultivar (S/O × Total).
Table 3: Optical density (OD) values in ELISA of diseased and healthy plants.
|
Varieties |
Sargodha |
Kot Momin |
Bhalwal |
|||
|
Diseased |
Healthy |
Diseased |
Healthy |
Diseased |
Healthy |
|
|
Citrus limetta |
0.82-2.22 |
0.18 |
0.89-2.35 |
0.17 |
0.85-2.32 |
0.13 |
|
C. reticulata cv Feutral’s early |
0.46-0.64 |
0.15 |
0.45-0.61 |
0.12 |
0.47-0.64 |
0.15 |
|
C. reticulata cv Kinnow |
0.25-0.56 |
0.13 |
0.28-0.53 |
0.11 |
0.26-0.52 |
0.12 |
The intensity of CTV infection was less in Tehsil Bhalwal and Sargodha as compared to Kot Momin among all 3 citrus cultivars. However, the infection was more in Mosambi followed by Feutrell’s early and Kinnow, respectively in both Tehsils. The range of OD values for infected samples of Mosambi in Tehsil Bhalwal was 0.85-2.32 while it was 0.82-2.22 in Sargodha. The healthy samples of Mosambi showed mean OD values of 0.13 and 0.18 values in Bhalwal and Sargodh, respectively. Feutrell’s early gave least OD values as compared to Mosambi in both Tehsils i.e., Bhalwal and Sargodha which range from 0.47-0.64 and 0.46-0.64, respectively. The healthy samples of Feutrell’s early gave 0.15 OD value in both Tehsils.
Disease incidence in three tehsils of Sargodha
The orchards of (Mosambi, Kinnow and Feutrell’s early) were surveyed based on symptomology in Sargodha, Bhalwal and Kotmomin to record disease incidence. The disease incidence was calculated based upon the salient CTV disease symptoms i.e., stem pitting and the abnormal development of root and scion stock union. Mosambi orchards were examined thoroughly for the detection of CTV based on symptomology. CTV showed the highest incidence rate of 91% in Kot Momin followed by 85% in Bhalwal and 64% in Sargodha (Figure 2). Similarly, symptomology-based detection was performed for 3 cultivars separately, among which Mosambi showed maximum disease incidence in all Tehsils followed by Feutrell’s early and Kinnow (Figure 3). The disease incidence of 3 citrus cultivars was recorded in following order i.e., Kot Momin, Bhalwal and Sargodha, respectively. The overall disease incidence and severity increased with every observation in all cultivars and Tehsils.
CTV disease severity in Mosambi orchards
Mosambi is one of the most affected citrus variety by the CTV therefore, the severity level of CTV was much higher in Mosambi plants. The increasing age of orchards showed more severe attacks of CTV by having the large number of pits in infected plants. It was estimated that the greater the age of the plant, the greater will be the severity of the virus. The maximum disease severity was recorded in age group 3 (11-15) years followed by age group 2 (6-10 years) and age group (up to 5 years), respectively (Figure 4).
Citrus tristeza virus (CTV) is one of the most important diseases, but it has not been given much importance in the past. It has spread fast in the country without being noticed. CTV is a huge threat
to the citrus industry all over the world as it is destroying the citrus orchard (Lin et al., 2006). CTV resulted in many disease epidemics worldwide and caused significant losses (Korkmaz et al., 2008). The citrus fields surveyed for sample collection and CTV was confirmed through ELISA. ELISA is the most efficient method of detecting the virus in plant cells amongst all methods of detecting virus. It is the cost effective, so it is commonly used in citrus growing countries (Hilf et al., 2005). The technique uses the capability of antibodies (which were extracted from animals) to recognize proteins, especially the viral coat protein. It is a quick and efficient method of detecting CTV and at a time large quantity of samples can be run (Folimonova, 2020). CTV was first reported in Pakistan in 1988 ona few citrus plants in KPK and Punjab and confirmed through ELISA (Bester et al., 2021). The suspected CTV samples were subjected to DAS-ELISA for the confirmation of virus. The intensity of yellow colour favours the intensity of the pathogen in the wells of the plate (Iftikhar et al., 2021). The wells of the plate with denser colour showed high OD values when placed in ELISA reader. A similar study for the detection of CTV was conducted by Iftikhar et al. (2009) and found the OD values at 405nm 0.60, 0.42 and 0.31 for sweet orange, Kinnow and grapefruit, respectively. The petioles and midribs were best tissues to detect CTV through DAS-ELISA (Anfoka et al., 2005). Wu et al. (2015) studied biological and serological characters of CTV for the better understanding of its dynamics. The darker colour of positive reaction is the indication of more virus titre as compared to light colours in the diseased samples of less affected plants. This study showed that maximum CTV disease incidence was recorded in Mosambi followed by Feutral’s early and Kinnow. Mosambi orchards were selected for the calculation of CTV disease severity being the more affected among others. Citrus limmetta (Mosambi) orchards showed increased disease severity in older orchards than younger ones. Previously, it was found by Sharma et al. (2011) that Citrus reticulata and Citrus limon were resistant against CTV disease. It is evident from all the studies that there was more disease incidence in sweet oranges. CTV disease incidence also varied on Kinnow with respect to different areas and it has serious role in citrus decline. In Florida, declined orchards were sampled randomly and subjected to high throughput sequencing (HTS) for the detection of exact cause and CTV was found (Britt et al., 2022). CTV disease was more severe in Sargodha than Bhalwal and Kot Momin. Naseem et al. (2016) recorded maximum CTV disease incidence in Sargodha (67%) followed by Bhalwal (48%) and Mardan (39%) collectively on all citrus cultivars. The variation in the level of incidence and severity may be the result of varied growing conditions and soil factors. The incidence of CTV disease depends upon the prevalence of aphid population (Harper et al., 2018). Arif et al. (2005) conducted extensive survey of citrus orchards and reported that percentage of infection was high in North western area of Pakistan which was 24-44%. CTV is a phloem limited virus where it replicates in sieve elements and interrupts the normal physiological functions of the citrus plants and also obstructs the translocation of prepared food (Britt et al., 2020). Moreno et al. (2008) revealed that (CTV) disease can be managed by using tristeza resistant rootstock, certifications programs of bud woods as well as through different quarantine measures. They observed that a complete study of the interaction between hosts its vectors as well as viral protein will provide a significant role for minimizing disease severity. The disease incidence and severity were increased with the passage of time, it is evident both from different dates of disease observation and varied age groups of the citrus cultivars in current study. Ippolito et al. (2020) used ordinary differential equations (ODEs) to describe the possible CTV disease outbreak with reference to time. He specified the joint likelihood function as poisson binomial distribution. Vidal et al. (2012) described that CTV incidence and severity increased in older plantations than in younger ones. The spatio-temporal distribution of CTV disease depends upon susceptibility of citrus cultivars, infectivity of virus isolates and population dynamics of vector (Moreno et al., 2008). The CTV disease severity was more in higher age groups which gradually decreased in lower age groups (Singh et al., 2017). In current study, CTV disease severity was more in Mosambi followed by Feutrell’s early and Kinnow, respectively. The research of Singh et al. (2017) endorsed the present study as they compared CTV disease incidence and severity in various citrus cultivars and found more in sweet oranges. In various countries, the negative impacts of CTV disease were countered by incorporating a cross-protection technique. Plants were inoculated with mild-strain of CTV to reduce the devastating effects of subsequent infections that can be introduced by aphids (Shilts et al., 2020).
Conclusions and Recommendations
The incidence and severity of CTV was high in sweet orange. CTV can be detected in three main parts of plants like twig, leaves and barks. This study provided a new quantitative information regarding spatial and temporal dynamics of CTV disease. Such data enabled the method of sampling designs were needed to quantify and compare the impact of climate, new management tactics and integrated disease management. The incidence and severity of CTV was more in Kot momin followed by Bhalwal and Sargodha. The old plantation is more in all these citrus growing areas. Therefore, it is concluded that CTV is well established in old plantations and the source of spread in new citrus plantations. Diagnosis of CTV in citrus nurseries and insect vector in Pakistan is the need of time.
Acknowledgments
The authors acknowledge the cooperation of the Plant Pathology Section, Ayub Agricultural Research Institute (AARI) Faisalabad (Pakistan) for conducting ELISA.
Novelty Statement
The spatial and temporal distribution of CTV disease based upon serological confirmation was studied for the first time in Sargodha, Pakistan.
Author’s Contribution
Muhammad Ahmad Zeshan and Yasir Iftikhar: Conceived the idea and supervised the research.
Asim Ali Khan and Maham Sohail: Conducted the experimentations.
Malik Abdul Rehman and Sonum Bashir: Helped in identification and collection of samples.
Muhammad Usman Ghani: Analyzed the data statistically.
Mustansar Mubeen: Refined and finalized the first draft.
Conflict of interest
The authors have declared no conflict of interest.
References
Abbas, M., M.M. Khan, B. Fatima, Y. Iftikhar, S.M. Mughal, M.J. Jaskani and H. Abbas. 2008. Elimination of Citrus tristeza closterovirus (CTV) and production of certified citrus plants through shoot-tip micro grafting. Pak. J. Bot., 40(3): 1301-1312.
Anfoka, G., M. Abhary, I. Fattash and M. Nakhla. 2005. Occurrence and distribution of citrus tristeza virus (CTV) in the Jordan Valley. Phytopath. Mediter., 44(1): 17-23.
Arif, M., A. Ahmad, M. Ibrahim and S. Hassan. 2005. Occurrence and distribution of virus and virus-like diseases of citrus in North-West Frontier province of Pakistan. Pak. J. Bot., 37(2): 407-410.
Bester, R., G. Cook and H.J. Maree. 2021. Citrus tristeza virus genotype detection using high-throughput sequencing. Viruses, 13: 168. https://doi.org/10.3390/v13020168
Britt, K., S. Gebben, A. Levy, D. Achor, P. Sieburth, K. Stevens, M. Al Rwahnih and O. Batuman. 2022. Analysis of citrus tristeza virus incidences within Asian citrus psyllid (Diaphorina citri) populations in florida via high-throughput sequencing. Insects, 13: 275. https://doi.org/10.3390/insects13030275
Britt, K., S. Gebben, A. Levy, M. Al Rwahnih and O. Batuman. 2020. The detection and surveillance of Asian citrus psyllid (Diaphorina citri) associated viruses in florida citrus groves. Front. Plant Sci., 10: 1687. https://doi.org/10.3389/fpls.2019.01687
Byamukama, E., D.L. Seifers, G.L. Hein, E. De Wolf, N.A. Tisserat, M.A.C. Langham, L.E. Osborne, A. Timmerman and S.N. Wegulo. 2013. Occurrence and distribution of Triticum mosaic virus in the central great plains. Plant Dis., 97: 21-29. https://doi.org/10.1094/PDIS-06-12-0535-RE
Folimonova, S.Y., 2020. Citrus tristeza virus: A large RNA virus with complex biology turned into a valuable tool for crop protection. PLoS Pathog., 16(4): e1008416. https://doi.org/10.1371/journal.ppat.1008416
Harper, S.J., S.J. Cowell and W.O. Dawson. 2018. Bottlenecks and complementation in the aphid transmission of citrus tristeza virus populations. Arch. Virol., 163(12): 3373-3376. https://doi.org/10.1007/s00705-018-4009-1
Hilf, M.E., V.A. Mavrodieva and S.M. Garnsey. 2005. Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology, 95: 909-917. https://doi.org/10.1094/PHYTO-95-0909
Hussain, Z., Y. Iftikhar, M. Mubeen, M.Z. Saleem, M.U. Naseer, M. Luqman, R. Anwar, F. Khadija and A. Abbas. 2021. Application of micronutrients enhances the quality of kinnow mandarin infected by citrus greening disease (Huanglongbing). Sarhad J. Agric., 38(1): 360-371. https://doi.org/10.17582/journal.sja/2022/38.1.360.371
Iftikhar, Y., M. Abbas, M. Mubeen, M. Zafar-ul-Hye, F. Bakhtawar, S. Bashir, A. Sajid and M.A. Shabbir. 2021. Overview of strain characterization in relation to serological and molecular detection of citrus tristeza clostero virus. Phyton Int. J. Exp. Bot., 90(4): 063-1074. https://doi.org/10.32604/phyton.2021.015508
. 2013. Symptomatic expression of Tristeza-infected citrus plants in Pakistan. Arch. Phytopath. Plant Protect., 46(1): 98-104. https://doi.org/10.1080/03235408.2012.734718
Iftikhar, Y., M.A. Khan, A. Rashid, S.M. Mughal, Z. Iqbal, A. Batool, M. Abbas, M.M. Khan, S. Muhammad and M. Jaskani. 2009. Occurrence and distribution of Citrus Tristeza Closterovirus in the Punjab and NWFP. Pak. J. Bot., 41(1): 373-380.
Ippolito, S., J. Laborde, T. Gottwald and M.S. Irey. 2020. Studying the spatial and temporal spread of citrus tristeza virus through ODEs and Bernoulli trials. J. Theoret. Biol., 497: 110279. https://doi.org/10.1016/j.jtbi.2020.110279
Korkmaz, S., B. Cevik, S. Önder and N.K. Koç. 2008. Biological, serological, and molecular characterization of citrus tristeza virus isolates from different citrus cultivation regions of Turkey. Turk. J. Agric. Forest, 32(5): 369-379.
Lin, Y., P.A. Rundell, L. Xie and C.A. Powel. 2006. Pre-reaction of citrus tristeza virus (CTV) specific antibodies and labelled secondary antibodies increases speed of direct tissue blot immunoassay for CTV. Plant Dis., 90: 675-679. https://doi.org/10.1094/PD-90-0675
Moreno, P., S. Ambrós, M.R. Albiach-Martí, J. Guerri and L. Pena. 2008. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Pathol., 9(2): 251-268. https://doi.org/10.1111/j.1364-3703.2007.00455.x
Naseem, S., S. Mahmood and Z. Ali. 2016. Occurrence of Citrus tristeza virus in Pakistan: A GIS based approach combining host distribution and disease reports. Pak. J. Agric. Sci., 53: 513-521. https://doi.org/10.21162/PAKJAS/16.3703
Sajid, A., M.U. Ghazanfar, S. Rauf, Z. Hussain, S. Ahmad and Y. Iftikhar. 2021. Incidence and molecular detection of greening disease in two citrus cultivars in Sargodha, Pakistan. Sarhad J. Agric., 37(1): 296-301. https://doi.org/10.17582/journal.sja/2021/37.1.296.301
Sharma S.K., A. Tarafdar, D. Khatun, S. Kumari and K.K. Biswas. 2011. Intra-farm diversity and evidence of genetic recombination of Citrus tristeza virus isolates in Delhi region of India. J. Plant Biochem. Biotech., 21: 38-43. https://doi.org/10.1007/s13562-011-0071-4
Singh, A.K., N.T. Meetei, B.K. Singh and N. Mandal. 2017. High incidence of citrus tristeza virus in mandarin (Citrus reticulata) in North-East states of India. Virus Dis., 28(4): 401-407. https://doi.org/10.1007/s13337-017-0411-7
Shilts, T., C.A. El-Mohtar, W.O. Dawson and N. Killiny. 2020. Citrus tristeza virus P33 protein is required for efficient transmission by the aphid Aphis citricidus. Viruses, 12: 1131. https://doi.org/10.3390/v12101131
Vidal, E., A. Moreno, E. Bertolini, M.C. Mart́ınez, A.R. Corrales and M. Cambra. 2012. Epidemiology of Citrus tristeza virus in nursery blocks of Citrus macrophylla and evaluation of control measures. Spanish J. Agric. Res., 10: 1107-1116. https://doi.org/10.5424/sjar/2012104-2813
Wu, G.W., M. Tang, G.P. Wang, F.Y. Jin, Z.K. Yang and L.J. Cheng. 2015. Genetic diversity and evolution of two capsid protein genes of citrus tristeza virus isolates from China. Arch. Virol., 160(3): 787-794. https://doi.org/10.1007/s00705-014-2281-2
To share on other social networks, click on any share button. What are these?








