A Thermally Stable Acidic Chitinase from Paenibacillus sp. Y412MC10: Molecular Characterization and its Structural Modeling
A Thermally Stable Acidic Chitinase from Paenibacillus sp. Y412MC10: Molecular Characterization and its Structural Modeling
Burarah Arooj1, Zeeshan Mutahir1, Moazzam Ali1, Mohsina Akhter2, Malik Siddique Mahmood1, Arslan Hamid3 and Mahjabeen Saleem1*
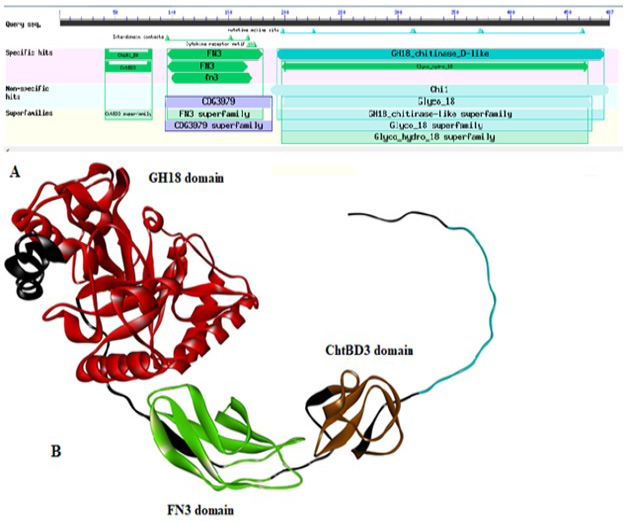
(A) Domain architecture of chitinase of Paenibacillus sp. Y412MC10 by using DbCAN. The locations of the following domains are indicated as: ChtBD3 (aa: 41-83); Fibronectin type III (FN3; aa: 97-173); GH18 (aa: 194-468). (B) 3D structure of chitinase enzyme showing its all domains in respective color.
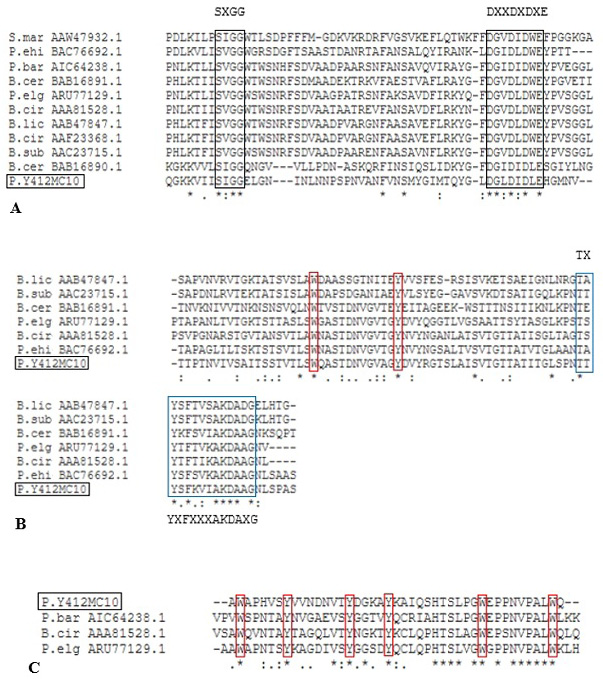
Alignment of putative CatD (A), Fn3D (B) ChBD3 (C) of Paenibacillus sp. Y412MC10 with different characterized bacterial chitinases. Black box indicates the conserved residues in GH 18 catalytic domain. Blue box indicates the conserved residues in Fn3 domain. Red box indicates the conserved aromatic residues W= Tryptophan and Y= Tyrosine, whereas, X may be any amino acid.
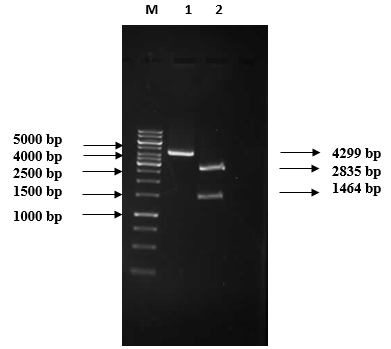
1% Agarose gel electrophoretic analysis of pRSETB/chitinase vector restriction digestion M: 1 kb DNA marker (Fermentas #SM0313); Lane 1: Recombinant plasmid (4299 bp); Lane 2: Double digested plasmid: 2835 bp pRSETB and 1464 bp chitinase gene.
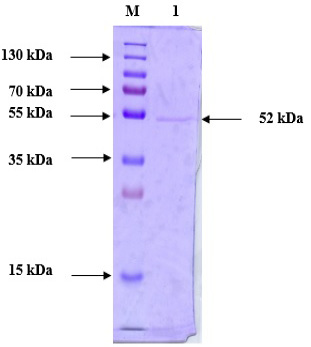
Purified protein SDS-PAGE analysis following anion exchange chromatography. Lane M: Prestained Protein Ladder (PageRuler™ #26616); Lane 1: Purified chitinase.
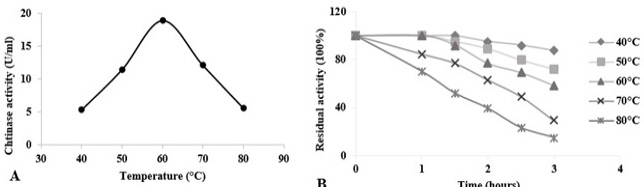
Temperature effects on chitinase activity and stability of Paenibacillus sp. Y412MC10 towards colloidal chitin at pH 5.5. (A) Assessment of optimal temperature for chitinase activity. (B) Evaluation of chitinase thermostability at temperatures from 40 °C to 80 °C. Activity of the non-heated enzyme was defined as 100%.
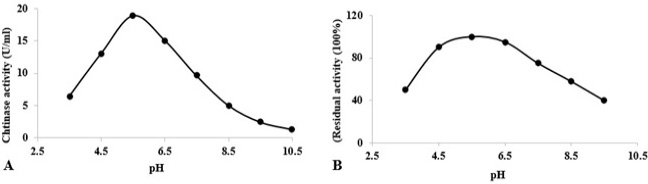
At varying pH values, stability and activity of chitinase towards colloidal chitin. (A) Determination of optimum pH for chitinase activity at 60°C. (B) Stability of chitinase after heating at different pH values for 1 h at 60°C. The activity of the enzyme after incubation at pH 5.5 (in acetate buffer) was defined as 100%.
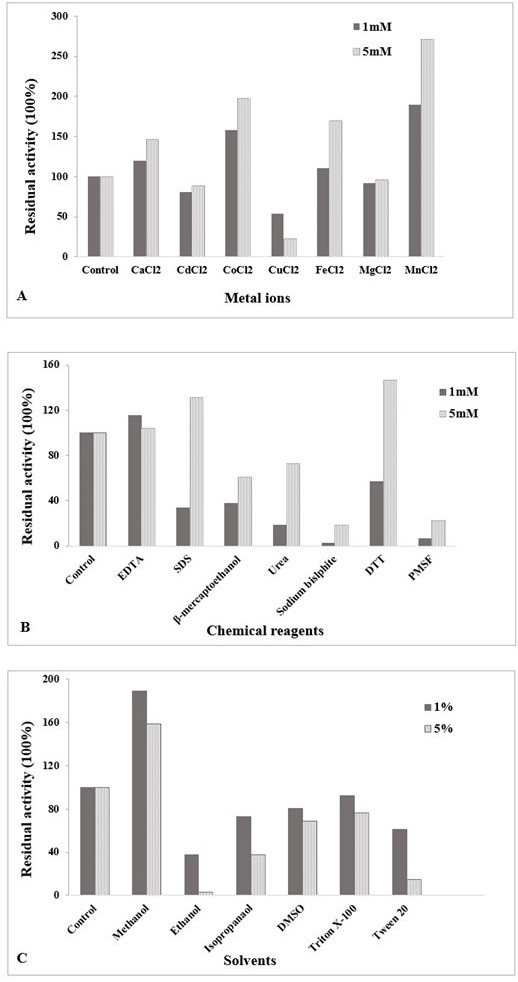
(A) Influence of metal ions on chitinase activity at different concentration (1 mM and 5 mM) of divalent cations. (B) Effect of chemical reagents on chitinase activity at 1 mM and 5 mM concentration. (C) Effect of solvents on chitinase activity at different percentages (1%, 5%). The enzyme was assayed at 60 °C and pH 5.5. Untreated enzyme was used as the control (defined as 100% activity). The data is representative of the mean of three experiments.
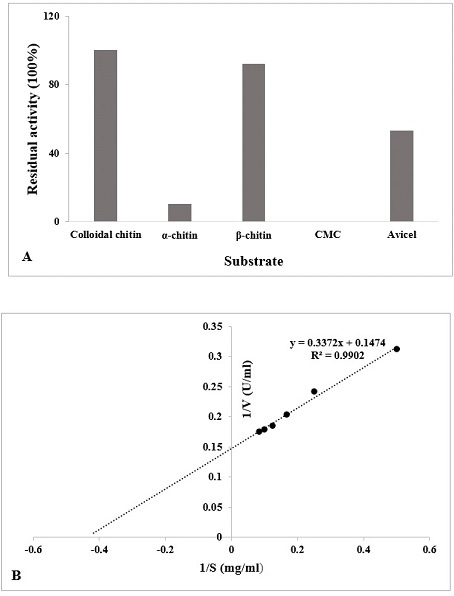
(A) Specificity of different substrate (1%) with the chitinase enzyme was determined (60°C, pH 5.5). The data is represented as % of activity relative to colloidal chitin. (B) Lineweaver-Burk double reciprocal plot of purified chitinase.
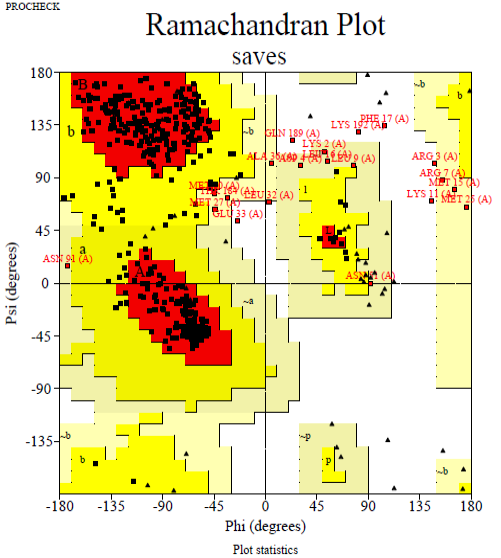
The Ramachandran plot of predicted chitinase indicating total residues in the the most favored areas (red region) and additional additional permitted areas regions (yellow region) are 82.9 and 12.3%, respectively.
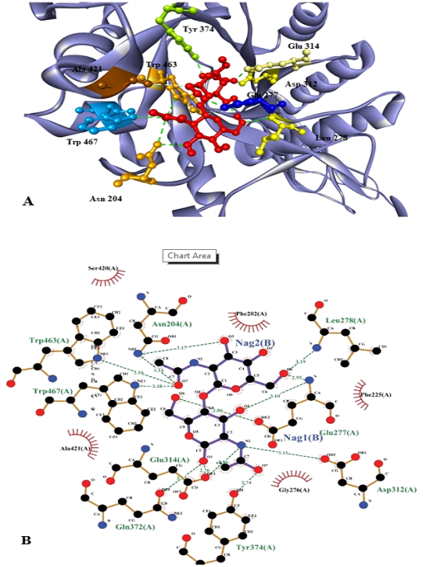
Substrate (NAG)2 docked with chitinase enzyme. Residues are named and the interactions are shown as green dotted lines. (A) 3D structure of the protein with substrate. Red molecules represents 2 molecules of NAG. (B) 2D structure of substrate (NAG)2 docked with chitinase enzyme which shows that active sites involve the interaction of 9 amino acids with 2 molecules of NAG.
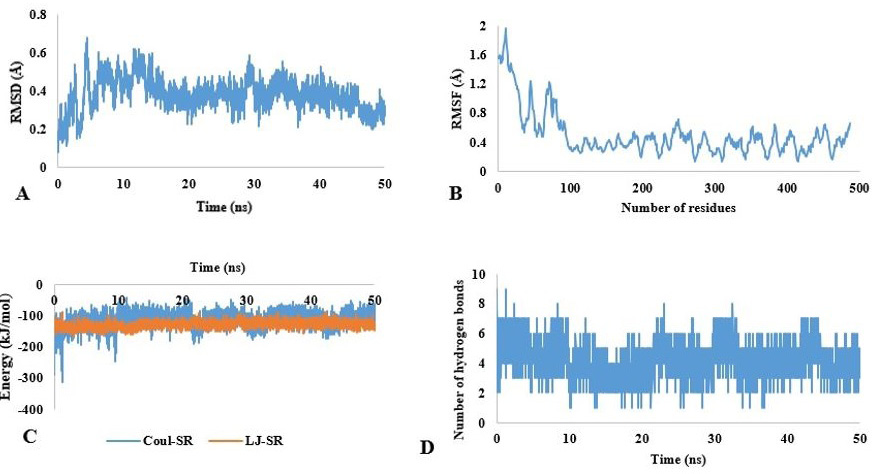
Molecular dynamic simulation of chitinase. (A) RMSD and (B) RMSF fluctuation values for chitinase. (C) Energy values of chitinase for 50 ns. (D) Hydrogen bonds (HB) depiction of chitinase-substrate complex.