Single Nucleotide Polymorphism in the Promoter Region of the IGF-1 Gene is Associated with Milk Production in Holstein and Jersey Cattle – Is the Aspect of Present Research Still Relevant in the Era of Genomic Selection?
Single Nucleotide Polymorphism in the Promoter Region of the IGF-1 Gene is Associated with Milk Production in Holstein and Jersey Cattle – Is the Aspect of Present Research Still Relevant in the Era of Genomic Selection?
Ewa Czerniawska-Piątkowska1, Iwona Szatkowska1, Daniel Zaborski2*, Wilhelm Grzesiak2, Sara Tabor-Osińska1, Małgorzata Wasielewska1,
Witold S. Proskura1, Wojciech Kruszyński3 and Edward Pawlina3
1Laboratory of Molecular Cytogenetics, Department of Ruminants Science, Faculty of Biotechnology and Animal Husbandry, West Pomeranian University of Technology, Szczecin, Klemensa Janickiego 29, 71-270 Szczecin, Poland
2Laboratory of Biostatistics, Department of Ruminants Science, Faculty of Biotechnology and Animal Husbandry, West Pomeranian University of Technology, Szczecin, Klemensa Janickiego 29, 71-270 Szczecin, Poland
3Department of Genetics, Faculty of Biology and Animal Breeding, Wrocław University of Environmental and Life Sciences, Kożuchowska 7, 51-631 Wrocław, Poland
ABSTRACT
The aim of the present study was to verify the potential effect of the C>T substitution at position -512 in the regulatory region of the IGF-1 gene on the level of milk performance traits in different cattle breeds. The study involved 227 Jersey, 147 Polish Holstein-Friesian black-and-white (HO) and 181 Polish Holstein-Friesian red-and-white (RW) cows. PCR-RFLP was used for genotyping. A bioinformatic analysis of the P1 regulatory sequence was also carried out. Three genotypes (TT, CT and CC) were identified. The CT genotype was the most frequent (0.40, 0.50 and 0.52 in Jerseys, HO and RW, respectively). The frequencies of the CC genotype were 0.22, 0.28 and 0.31 in the RW, Jersey and HO breeds, respectively, whereas those of the TT genotype were 0.19, 0.26, and 0.32 for HO, RW, and Jerseys, respectively. For Jerseys, significant differences in milk yield, fat and protein percentage were found in the second lactation. In HO, significant differences in milk fat content and yield were observed in the third lactation. No significant differences were found for RW. The bioinformatic analysis allowed us to infer that transcription factors other than the ZFP217 protein bind to the sites located outside the C>T substitution. Further studies are required to elucidate the molecular basis of the relationships observed in the present research.
Article Information
Received 08 December 2019
Revised 02 February 2020
Accepted 20 February 2020
Available online 08 October 2021
Authors’ Contribution
ECP, IS, WG, WK and EP conceived and designed the study. WSP, DZ and MW collected the data. WSP, DZ and STO analyzed and interpreted the data. WSP, DZ, STO, MW and WK drafted the article. ECP, IS, WG and EP critically reviewed the article.
Key words
Insulin-like growth factor 1, Polymorphism, Jersey, Holstein-friesian, Milk yield
DOI: https://dx.doi.org/10.17582/journal.pjz/20191208211212
* Corresponding author: [email protected]
0030-9923/2021/0006-2295 $ 9.00/0
Copyright 2021 Zoological Society of Pakistan
INTRODUCTION
An interest in insulin-like growth factor 1 (IGF-1) in terms of the potential creation of variability in milk performance traits in cattle results from several premises. The most important ones include: 1) the localization of the gene coding for this factor in the QTL region for dairy and beef traits (BTA5) (Smaragdov et al., 2006) and 2) its pleiotropic action on many tissues and organs (including the mammary gland) consisting of various cell types, at different stages of growth, differentiation and secretion activity (Connor et al., 2007).
A mature IGF-1 molecule, highly conservative in mammals, is a basic peptide of 70 amino acids, which, in principle, has an endocrine action, but can exert a paracrine action as a locally synthetized growth factor (Dai et al., 2010). In the bloodstream and extracellular environment, IGF-1 forms complexes with binding proteins (IGFBP 1 – 6), mainly with IGFBP-3 and the acid-labile (alpha) subunit (ALS), which modulate its bioavailability. In a form free from such complexes, it acts on target cells through a specific membrane receptor (IGF-1R). The binding of a ligand to its receptor activates a tyrosine kinase and subsequently the phosphorylation of the IRS-1 protein, thereby triggering a cascade of reactions of signaling molecules, through which the signal is transduced to the cell nucleus. In this manner, the transcription of target genes occurs. Liver hepatocytes are the major site of IGF-1 synthesis in response to growth hormone (GH). It can also be locally synthetized in many tissues in response to a wide spectrum of specific transcription factors. IGF-1 has a wide range of biological activity. It plays a significant role in pre- and postnatal development and affects anabolism and tissue repair processes in adults. It is also a natural mitogen and stimulator of cell growth and differentiation, as well as an apoptosis inhibitor. In addition, it stimulates DNA, RNA and protein synthesis, which has been confirmed in in vitro cultures (Connor et al., 2007; Bartke et al., 2016; Hellström et al., 2016).
Despite the simple structure of the IGF-1 molecule, the structure and regulation of IGF-1 gene expression are exceptionally complex and its mechanism is highly conservative in mammals. The bovine IGF-1 gene, which was mapped to chromosome 5 (Bishop et al., 1991), consists of six exons interspersed with five introns. The mature IGF-1 molecule is encoded by exons 3 and 4 (Wang et al., 2003), whereas exons 5 and 6 code for an alternative E domain, whose function has not yet been determined. Two promoters (P1 and P2), controlling two leader exons (1 and 2, respectively), are involved in the regulation of IGF-1 gene transcription. As a result of alternative splicing, expression control through two promoters, and numerous transcription start sites, many different forms of IGF-1 mRNA are synthetized, which are generally known as class 1 or 2 transcripts. What is interesting is the fact that class 1 IGF-1 mRNA predominates in cattle, with the highest expression level in the liver, adipose tissue, male gonads, spleen and mammary gland. Moreover, it has been shown that the translation of class 1 IGF-1 mRNA occurs with a four times higher efficiency compared with class 2 IGF-1 mRNA, although some tissue-specific differences exist (Wang et al., 2003).
In the above context, changes in the regulatory sequence of the IGF-1 gene may play a significant role in the IGF-1 protein level. In cattle, several polymorphic sites have been identified in this region, which may potentially affect IGF-1 expression level. Chung et al. (2015) described a C/A substitution at position -323, Curi et al. (2005) identified a CA(10-11) microsatellite polymorphism at positions -326 – 349 and Ge et al. (2001) reported a C/T transition at position -512. Especially the last one was an object of interest in terms of its potential effect on the IGF-1 gene expression level, since some relationships between beef performance and the discussed polymorphism have been found, while the results for milk traits are ambiguous (Siadkowska et al., 2006; Szewczuk et al., 2012).
Therefore, the aim of the present study was to verify the hypothesis about the potential effect of the afore-mentioned substitution on the level of milk traits in different cattle breeds.
MATERIALS AND METHODS
Animals
The study involved a total of 227 Jersey, 147 Polish Holstein-Friesian black-and-white (HO) and 181 Polish Holstein-Friesian red-and-white (RW) cows from three herds located in West Pomerania, Opole and Grater Poland Provinces.
Feeding
Feeding was based on a total mixed ration (TMR), mainly composed of maize silage, grass haylage, maize cereals, oat cereals, soybean meals and mineral-vitamin mixtures.
DNA isolation and genotyping
DNA was isolated with the MasterPureTM Genomic DNA Purification Kit (Epicentre Biotechnologies). DNA extractions were stored at -20°C for further analysis.
The PCR reaction contained approximately ~50 ng of the DNA template, 0.3 units of DreamTaq DNA Polymerase, 1×PCR Buffer, 1.5 mM MgCl2, 200µM each dNTP, 15 pmol of each primer and filled up to 15 µL with deionized water. The polymorphic site (rs109763947) was described by Ge et al. (1997). The C→T transition (GenBank Acc. No. AF017143) is located in the promoter region of the bovine IGF-1 gene at 512 bp 5’ to the first codon of the first exon (Ge et al., 2001). The following primer sequences designed by Ge et al. (2001) were used: IGF677F, 5′-ATTACAAAGCTGCCTGCCCC-3′ and: IGF897R, 5′-ACCTTACCCGTATGAAAGGAATATACGT-3′.
The thermocycler conditions were as follows: denaturation at 96°C/2 min, followed by 31 cycles at 94°C/60 s, primer annealing at 62°C/45 s, amplicon synthesis at 72°C/60 s and final synthesis at 72°C/5 min. The specificity and efficiency of the amplification reaction (4 μl) were verified by electrophoresis on 1.5% agarose gels (Syngen) in 1×TBE. The PCR products (11µl) were digested for 3 h at 37°C with two units of Eco105I (SnaBI) (ThermoScientificTM) restriction endonuclease (NEB) recognizing the TAC↓GTA sites. The digestion reaction contained also 2 µL 10×Tango buffer and H2O up to a total volume of 20 µL. After incubation, the digested fragments were separated on a 3% agarose gel stained with ethidium bromide in 1×TBE for 50 min at 130 V and visualized under UV light.
Statistical analysis
Statistical analysis was performed using R software (R Core Team, 2015). An additive relationship matrix was calculated based on a three-generation pedigree using the kinship 2 R package (Sinnwell et al., 2014). The following linear model was estimated using the lmekin function of the coxme R package (Therneau, 2018):
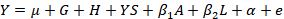
Where, Y is the phenotypic value of each trait, µ is the overall mean, G is the fixed effect of genotype, H is the fixed effect of herd, YS is the fixed effect of year-season of calving, β1A is the regression coefficient for age of cow, β2L is the regression coefficient for lactation length, α is a random polygenic component accounting for all known pedigree relationships, and e is a random residual.
In the analyses performed simultaneously for all three lactations, the fixed effect of lactation was also included. The Bonferroni correction was applied for multiple comparisons.
Bioinformatic analysis
A bioinformatic analysis of the P1 regulatory sequence of the bovine IGF-1 gene (GenBank Acc. No. AF210383.1) was carried out using the Softberry (Softberry, Inc., Mount Kisco, NY, USA) and Tfsitescan (the Institute of Transcriptional Informatics, Pittsburgh, PA, USA) programs. The analysis was aimed at predicting potential transcription factor binding consensus sites within or in close proximity to the investigated substitution (c.-512 C>T).
RESULTS
Genotype and allele frequency
In the study herds, three genotypes (TT, CT and CC) corresponding to the transition (c. -512 C>T) in IGF-1 determined using the SnaBI restrictase were identified. The 249-bp PCR product was characteristic of the CC genotype, whereas the TT genotype was identified as a 223-bp band after digestion with the restriction endonuclease. The 26-bp restriction fragment was undetectable (Fig. 1).
The largest group consisted of individuals carrying the heterozygous CT genotype (two bands) in all study herds and its frequency was 0.40, 0.50 and 0.52 in Jerseys, Polish Holstein-Friesian black-and-whites and Polish Holstein-Friesian red-and-whites, respectively (Table I). The highest frequency of the CC genotype (0.31) was observed in the herd of Polish Holstein-Friesian black-and-white cows and the lowest one (0.22) in that of Polish Holstein-Friesian red-and-white cows, with the intermediate values (0.28) in Jerseys. The frequency of the TT genotype was also diverse: the highest (0.32) was found in Jersey individuals and the lowest (0.19) in Polish Holstein-Friesian black-and-white cows. The allele frequency in individual herds was similar. The T allele predominated in the herd of Polish Holstein-Friesian black-and-white cows (0.56), with its lowest frequency in Polish Holstein-Friesian red-and-white and Jersey cows (0.44). The frequency of the C allele was opposite: 0.44 in Polish Holstein-Friesian black-and-whites and 0.48 in Polish Holstein-Friesian red-and-whites and Jerseys.
Table I. Genotype and allele frequencies for IGF1/SnaBI in the study herds.
|
Genotypes |
Alleles |
||||
|
TT |
CT |
CC |
T |
C |
|
|
Jersey |
|||||
|
n = 227 |
73 |
91 |
63 |
0.52 |
0.48 |
|
Frequency |
0.32 |
0.40 |
0.28 |
||
|
Polish Holstein-Friesian black-and-white |
|||||
|
n = 147 |
27 |
74 |
46 |
0.56 |
0.44 |
|
Frequency |
0.19 |
0.50 |
0.31 |
||
|
Polish Holstein-Friesian red-and-white |
|||||
|
n = 181 |
47 |
94 |
40 |
0.52 |
0.48 |
|
Frequency |
0.26 |
0.52 |
0.22 |
||
An association between milk production traits and the IGF1/SnaBI genotypes
The values of milk performance traits for three 305-day lactations in the herd of Jersey cows depending on the IGF1/SnaBI genotype are presented in Table II. In the first lactation, in which an average milk yield was 5703 kg, no statistically significant differences were found. Nevertheless, the homozygous CC cows had a 342 kg higher milk yield than the TT homozygotes. Heterozygotes were characterized by an intermediate milk yield. The CC cows also had a somewhat higher milk protein content and yield as well as milk fat yield at a lower fat content. Statistically significant differences in selected traits were found in the second lactation. The milk yield of the homozygous CC cows was significantly higher (+444 kg; p=0.0442) than that of the TT homozygotes. The effect of the C allele in the heterozygous CT configuration, although not statistically significant, was also noticeable, which resulted in a 301 kg higher milk yield in comparison with the TT cows. On the other hand, milk fat concentration in the TT individuals was significantly higher (p=0.0067) compared with the CC homozygotes (+0.44%) and the CT heterozygotes (+0.28%). Similar, statistically significant differences (p=0.0263) were observed in milk protein content (3.99%, 3.88%, and 3.89% for the TT, CT and CC genotypes, respectively). However, this was not reflected in the total milk protein yield for the whole 305-day lactation period in the TT homozygotes, which was greater (+10 kg) in the CC cows with a markedly higher milk yield, although the difference was not statistically significant. In the third lactation (similarly to the first one), no significant differences in the milk yield of cows carrying different IGF1/SnaBI genotypes were found. However, it is worth mentioning that the heterozygous CT individuals were characterized by the highest milk yield in this lactation, which has not been previously observed. On the other hand, a trend regarding the superiority of the TT cows over the CT and CC individuals in terms of milk protein and fat percentage, which was noticed in the second lactation, was also confirmed in the third lactation.
The values of milk performance traits for three 305-day lactations in the herd of Polish Holstein-Friesian black-and-white cows depending on the IGF1/SnaBI genotype are presented in Table III. In all three lactations, the highest milk yield was characteristic of the homozygous CC individuals compared with the TT ones, which was not confirmed statistically but was increasingly more pronounced with each successive lactation (+141 kg, +185 kg, and +367 kg in the first, second and third lactation, respectively). Heterozygotes were characterized by the intermediate values of this trait. Milk fat content and yield in the third lactation were the only traits with significant differences among genotypes. What is interesting is that the lowest milk fat percentage was determined in the milk from the heterozygous CT cows compared with the CC homozygotes (p=0.0181). This difference amounted to 0.17%. The level of this trait in the TT homozygotes was similar to that in the CC ones. A high milk fat content and the highest milk yield of the CC cows resulted in the highest fat yield in the carriers of this genotype, which was confirmed statistically (p=0.0475). This difference was +27 kg in comparison with the CT individuals and +21 kg with the TT homozygotes. When summarizing performance data presented in Table III, it can be noticed that all the investigated traits (with significant and non-significant differences), except for milk protein percentage, had the most favorable values in the CC individuals.
The values of milk performance traits for three 305-day lactations in the herd of Polish Holstein-Friesian red-and-white cows depending on the IGF1/SnaBI genotype are presented in Table IV. In contrast to the aforementioned breeds, no significant differences in milk yield, milk protein and fat content or yield among individuals of different genotypes were found in the herd of Polish Holstein-Friesian red-and-white cows. However, a repetitive trend of the higher milk yield of cows with the homozygous CC genotype compared with the heterozygotes (+291 kg) and the TT homozygotes (+144 kg) was noticeable in the first two lactations. On the other hand, the highest values of this trait in the third lactation were found in heterozygotes and were 701 kg greater than those in the CC individuals and 650 kg greater than those in the TT homozygotes. Such relationships were also observed in the third lactation of Jersey cows. In all studied lactations, Polish Holstein-Friesian red-and-white cows carrying the CC genotype produced milk with the lowest fat content. The most favorable values of this trait were recorded in the CT heterozygotes, except for the second lactation, in which a slight superiority of the TT homozygotes (0.03%) was recorded. Finally, it should be emphasized that the IGF1/SnaBI genotypes affected the milk protein and fat content in Polish Holstein-Friesian red-and-white cows only to a minimal extent.
Bioinformatic analysis results
Using available databases and verifying potential transcription factor binding sites reported by other authors through the comparison of the consensus for the indicated regulatory proteins, it can be assumed (with a high probability) that other transcription factors (besides the ZFP217 protein) bind to the sites located outside the investigated C>T substitution at position -512 (Table VI).
DISCUSSION
The allele and genotype frequencies obtained in the present study in the herds of Jersey, Polish Holstein-Friesian black-and-white and Polish Holstein-Friesian red-and-white cows were similar, although the frequency of the C allele was somewhat higher in the first and last of the aforementioned breeds in comparison with Polish Holstein-Friesian black-and-whites. When comparing the results of the present study with those of other authors (Table V), it is worth mentioning that, with few exceptions, the T allele slightly predominated in the herds of typical dairy breeds, whereas the C allele was more frequent in the herds of beef breeds. However, these differences were not statistically significant.
Table II. Milk production traits in association with the IGF1/SnaBI genotype of Jersey cows.
|
Lactation |
Trait |
Total |
Genotype |
P |
||
|
CC |
CT |
TT |
||||
|
I |
MY [kg] |
5703.5±913.28 |
5902.9±934.91 |
5680.2±948.26 |
5560.47±827.77 |
0.1329 |
|
FY [kg] |
289.87±43.1 |
293.05±40.66 |
291.49±46.19 |
285.1±41.3 |
0.8900 |
|
|
F [%] |
5.11±0.54 |
5.01±0.57 |
5.16±0.52 |
5.14±0.54 |
0.1223 |
|
|
PY [kg] |
218.39±33.4 |
226.98±33.73 |
217±34.2 |
212.7±30.97 |
0.0686 |
|
|
P [%] |
3.87±0.27 |
3.89±0.32 |
3.86±0.25 |
3.85±0.23 |
0.9885 |
|
|
II |
MY [kg] |
6451.75±938.64 |
6638.41±1126.62a |
6495.61±829.81 |
6194.77±824.08b |
0.0442 |
|
FY [kg] |
332.84±47.36 |
328.32±45.87 |
333.59±46.36 |
336.56±50.99 |
0.8544 |
|
|
F [%] |
5.19±0.57 |
5±0.55a |
5.16±0.54a |
5.44±0.54b |
0.0067 |
|
|
PY [kg] |
252.48±35.62 |
256.85±40.82 |
253.19±31.31 |
246.9±35.6 |
0.2559 |
|
|
P [%] |
3.91±0.24 |
3.89±0.22a |
3.88±0.23a |
3.99±0.26b |
0.0263 |
|
|
III |
MY [kg] |
6478.62±1059.56 |
6399.91±1216.13 |
6693.39±1082.62 |
6281.56±858.48 |
0.4689 |
|
FY [kg] |
336.36±61.12 |
319.18±62.5 |
352.94±59.86 |
330.92±58.59 |
0.3550 |
|
|
F [%] |
5.21±0.54 |
4.96±0.62 |
5.29±0.41 |
5.34±0.54 |
0.2993 |
|
|
PY [kg] |
254.35±43.4 |
247.55±46.92 |
260.74±44.61 |
252.4±39.12 |
0.8499 |
|
|
P [%] |
3.93±0.24 |
3.87±0.28 |
3.9±0.22 |
4.02±0.19 |
0.1905 |
MY, milk yield; FY, fat yield; F, fat content; PY, protein yield; P, protein content, a, b, different superscript letters within rows denote statistical significance at P≤0.05
Table III. Milk production traits in association with the IGF1/SnaBI genotype of Polish Holstein-Friesian black-and-white cows.
|
Lactation |
Trait |
Total |
Genotype |
P |
||
|
CC |
CT |
TT |
||||
|
I |
MY [kg] |
10077.06±1554.47 |
10222.39±1528.93 |
10016.78±1559.74 |
10081.63±1571.05 |
0.5322 |
|
FY [kg] |
345.56±57.77 |
356.89±59.87 |
344.15±55.15 |
338.94±60.53 |
0.1623 |
|
|
F [%] |
3.46±0.51 |
3.52±0.51 |
3.47±0.54 |
3.38±0.45 |
0.4328 |
|
|
PY [kg] |
329.68±48.52 |
332.97±47.71 |
328.41±48.67 |
329.59±49.23 |
0.7853 |
|
|
P [%] |
3.28±0.17 |
3.26±0.18 |
3.29±0.17 |
3.28±0.16 |
0.3518 |
|
|
II |
MY [kg] |
10566.66±1858.49 |
10663.05±2109.95 |
10569.16±1731.62 |
10478.99±1909.04 |
0.5533 |
|
FY [kg] |
379.58±74.46 |
383.35±71.68 |
380.22±75.77 |
374.99±74.56 |
0.5795 |
|
|
F [%] |
3.61±0.5 |
3.64±0.49 |
3.61±0.5 |
3.6±0.52 |
0.8509 |
|
|
PY [kg] |
348.73±58.18 |
351.04±62.58 |
349.4±55.18 |
345.34±61.01 |
0.4765 |
|
|
P [%] |
3.31±0.19 |
3.31±0.22 |
3.31±0.18 |
3.3±0.19 |
0.6452 |
|
|
III |
MY [kg] |
10625.47±2198.49 |
10812.41±2235.26 |
10651.61±2301.76 |
10445.83±1966.08 |
0.1432 |
|
FY [kg] |
384.94±81.49 |
405.13±87.18a |
378.51±82.67b |
384.32±73.79b |
0.0475 |
|
|
F [%] |
3.65±0.47 |
3.76±0.43a |
3.59±0.49b |
3.7±0.43 |
0.0181 |
|
|
PY [kg] |
346.97±69.51 |
351.25±69.57 |
347.84±72.5 |
342.31±63.81 |
0.1468 |
|
|
P [%] |
3.28±0.2 |
3.26±0.18 |
3.28±0.2 |
3.28±0.21 |
0.8970 |
MY, milk yield; FY, fat yield; F, fat content; PY, protein yield; P, protein content, a, b, different superscript letters within rows denote statistical significance at P≤0.05
Table IV. Milk production traits in association with the IGF1/SnaBI genotype of Polish Holstein-Friesian red-and-white cows.
|
Lactation |
Trait |
Total |
Genotype |
P |
||
|
CC |
CT |
TT |
||||
|
I |
MY [kg] |
9527.21±2043.08 |
9719.07±1668.27 |
9428.41±2095.23 |
9575.35±2241.78 |
0.9812 |
|
FY [kg] |
359.2±71.89 |
363.34±61.13 |
359.2±76.46 |
355.9±72.02 |
0.7654 |
|
|
F [%] |
3.82±0.55 |
3.77±0.48 |
3.86±0.57 |
3.79±0.59 |
0.6557 |
|
|
PY [kg] |
324.17±64.19 |
328.59±57.12 |
321.02±66.45 |
327.05±66.26 |
0.9269 |
|
|
P [%] |
3.42±0.22 |
3.39±0.23 |
3.42±0.21 |
3.45±0.25 |
0.5056 |
|
|
II |
MY [kg] |
11037.82±2105.88 |
11584.38±1457.59 |
10753.06±2311.03 |
11465.81±1804.27 |
0.2327 |
|
FY [kg] |
415.78±82.09 |
427.99±71.12 |
406.13±88.44 |
435.43±68.35 |
0.5865 |
|
|
F [%] |
3.81±0.59 |
3.7±0.48 |
3.82±0.62 |
3.85±0.61 |
0.8347 |
|
|
PY [kg] |
374.88±65.38 |
388.25±41.92 |
365.66±72.12 |
392.26±55.99 |
0.2009 |
|
|
P [%] |
3.41±0.22 |
3.36±0.14 |
3.42±0.23 |
3.44±0.26 |
0.6230 |
|
|
III |
MY [kg] |
11374.23±2500.7 |
10878.25±3613.03 |
11579.44±2638.73 |
10929.63±1400.63 |
0.5501 |
|
FY [kg] |
441.92±105.65 |
391.88±127.41 |
454.26±103.83 |
425.31±106.12 |
0.2539 |
|
|
F [%] |
3.9±0.53 |
3.62±0.29 |
3.95±0.49 |
3.87±0.73 |
0.5894 |
|
|
PY [kg] |
383.57±82.47 |
363.69±116.94 |
390.92±87.97 |
368.69±40.4 |
0.4561 |
|
|
P [%] |
3.38±0.21 |
3.36±0.08 |
3.38±0.22 |
3.39±0.22 |
0.6629 |
MY, milk yield; FY, fat yield; F, fat content; PY, protein yield; P, protein content.
Table V. The frequency of the T and C alleles reported by other authors.
|
Breed |
Allele C |
Allele T |
Author |
|
Beef breeds |
|||
|
Angus |
0.36 |
0.64 |
|
|
Charolais |
0.74 |
0.26 |
|
|
South Anatolian Red |
0.77 |
0.23 |
|
|
East Anatolian Red |
0.63 |
0.38 |
|
|
Angus |
0.56 |
0.44 |
|
|
Dairy breeds |
|||
|
HF* black-and-white Dairy traits** |
0.52 |
0.48 |
|
|
HF black-and-white Dairy traits |
0.52 |
0.46 |
|
|
HF black-and-white Beef traits*** |
0.40 |
0.60 |
|
|
HF black-and-white Beef traits |
0.41 |
0.59 |
|
|
HF black-and-white Beef traits |
0.44 |
0.56 |
|
|
HF black-and-white |
0.43 |
0.56 |
Our data |
|
Jersey |
0.47 |
0.53 |
|
|
HF red-and-white |
0.48 |
0.52 |
|
*HF, Holstein-Friesian; **Dairy traits, dairy traits analysis; ***Beef traits, beef traits analysis.
In the past, attempts have been made to associate the C>T substitution at position -512 in the regulatory region of the IGF-1 gene mainly with the dressing percentage in beef cattle. And so, Ge et al. (2001) like Siadkowska et al. (2006) and De la Rosa Reyna et al. (2010) found significant associations between the CC genotype and the higher body weight of cows at an older age. What is significant is that the CC genotype was positively correlated with the meat and fat weight of the carcass (Siadkowska et al., 2006). On the other hand, Akis et al. (2010) reported that the IGF-1/SnaBI genotypic effect was hardly noticeable in primitive breeds, characterized by the low indices of meat performance traits. Mullen et al. (2011) and De la Rosa Reyna et al. (2010) hypothesized that the association between the aforementioned transition and phenotype involved mature individuals of beef breeds rather than the body weight of younger animals. To a lesser extent, research interest has been focused on the analysis of milk performance traits in the context of their association with the aforementioned substitution in the regulatory region of the IGF-1 gene, and no statistically significant relationships were found between them (Siadkowska et al., 2006; Szewczuk et al., 2012). It is, however, noticeable that the highest yields were recorded in the CT heterozygotes from both above-mentioned herds. This relationship was observed for total milk yield and protein and fat content. In the present study, statistically significant differences were found in the groups of cows carrying different IGF-1/SnaBI genotypes. Nevertheless, they did not reflect all the analyzed lactations. However, if one takes into account the combined milk yield of individuals with different genotypes for all lactations, then the highest yields were recorded in the CC homozygotes, irrespective of dairy cattle breeds that differ not only in milk yield but also fat and protein content.
Table VI. A partial sequence of the regulatory region of the bovine IGF-1 gene with the site of the C>T substitution at position -512 (GenBank).
In the above context, it would be of interest to determine whether and in which way the C>T substitution at position -512 in the regulatory region of the IGF-1 gene affects the level of IGF-1 synthesis and therefore the phenotypic traits controlled by this protein. It seems justified to assume that the described polymorphism may create or remove potential consensus sites for transcription factors, which could not be confirmed in the present study. However, based on previously published results, a selected sequence of the regulatory region of the bovine IGF-1 gene is presented, taking into account the substitution site and the sequences of potential consensus sites for transcription factors indicated by other authors. And so, Mullen et al. (2011), reporting the higher body weight of the adult CC individuals, showed at the same time that the allele with cytosine introduced two new binding sites for the HSF1 and ZFP 217 factors. It is, however, noticeable that, in the case of HSF1, the binding site is located outside the described substitution, and the consensus for ZFP 217 is not complete (CAGAA). Although the sequence of the latter has not yet been determined in cattle, it can be assumed (with a high probability) that both aforementioned proteins act as repressors and not activators of transcription. Therefore, it would be difficult to explain a more favorable phenotype of the CC cows based on the above information. On the other hand, Islam et al. (2009), investigating a population of beef cattle, suggested that the C allele probably introduces a new binding site for the NF1 transcription factor, commonly known as an activator or repressor of many target genes (Gronostajski, 2000). Hence, it is worth focusing on the function of this factor in adipose tissue, with which the values of phenotypic traits have been correlated by the aforementioned authors, indicating a statistically higher fat content in the CC individuals.
And so, NF1 initiates a high gene expression involved in the process of preadipocyte differentiation into mature cells of white adipose tissue (adipocytes). It should be emphasized that these processes are accompanied by an increased expression of IGF-1 (Islam et al., 2009). Consequently, a higher amount of adipose tissue in cows carrying the CC genotype can be explained assuming that the C>T substitution indeed introduces a consensus site for NF1. However, the consensus for this protein is not complete, which makes the above considerations about NF1 speculative (Nagata et al., 1983; Nowock et al., 1985; Gronostajski, 2000; Miura et al., 2004).
Irrespective of the action of the molecular mechanism of the C>T substitution at position -512 of the bovine IGF-1 gene, the research on the level of IGF-1 gene expression in the liver and the concentration of the IGF-1 protein in the blood of cows with different genotypes of the described substitution carried out by Maj et al. (2008), is noteworthy. And so, the highest expression of IGF-1 was noticed in cows with the CC genotype, compared with CT and TT. Also, the concentration of the IGF-1 protein was statistically significantly different. It was highest in the CC individuals (1024 ng/ml) and lower in the CT (859 ng/ml) and TT (698 ng/ml) ones. The cited authors also indicated a consensus site for NF1 near the substitution site in the in silico analysis. Similar relationships, except for the in silico analysis, were reported by Ruprechter et al. (2011). Therefore, it can be stated with a high probability that the investigated P1 promoter region of the bovine IGF-1 gene modulates its expression level, whose main site is the liver (possibly not only in response to GH). Assuming the above, it should also be stated that the principal effect of IGF-1 on the functions of the mammary gland in cows of the different C/T genotypes probably has an endocrine character, although a paracrine action cannot be excluded.
In the above context, it is worth considering the role of IGF-1 in the mammary gland at these two levels (endo- and paracrine), although according to Murney et al. (2015), the nature of the dynamics of the IGF-1 biochemical changes in this gland has not been completely understood. However, a high secretion of the local IGF-1 in the mammary gland was observed in heifers and primiparae at the first stages of gestation (from 194 to 213 days), i.e. during the intensive development of this organ (Plath-Gabler et al., 2001). As a powerful mitogen, it is involved in the proliferation of epithelial cells at this time, which was confirmed under both in vivo and in vitro conditions. It also protects cells against apoptosis (Akers et al., 2000). A high secretion of IGF-1 still remained at a relatively high level during lactogenesis but it rapidly declined at lactation peak in order to increase again during involution (Plath-Gabler et al., 2001). Murney et al. (2015) suggests that the low level of the local IGF-1 in the mammary gland during lactopoiesis indicates that this is the endocrine action of IGF-1 that is important at this stage. But is it significant at this time? Although it has not been observed in cows, but goats that received IGF-1 in the form of intra-arterial injections were characterized by an increased milk secretion (Murney et al., 2015). Therefore, if we assume its important functions during lactopoiesis and compare the results obtained by Maj et al. (2008) with these suggestions, the higher yields of cows with the CC and CT genotypes compared with the TT ones could be explained, even if the differences were observed only in some lactations.
CONCLUSIONS
Finally, one should attempt to answer the question included in the title of the present study. An identification of the polymorphisms in the coding, non-coding and regulatory sequences of genes in association with the level of production traits in cattle is a very significant stage of marker-assisted selection (MAS), although it should be emphasized that not all described SNP allowed for drawing definite conclusions. Genomic selection offers great opportunities, since it is based on the comprehensive use of knowledge of the identified SNP in the form of predictive equations, which enable the assignment of breeding value to them or their haplotypes. In the above context, an identification of new markers or a verification of the already known ones at the biological and production levels may still be important for the essence of genomic research.
Statement of conflict of interest
The authors have declared no conflict of interest.
References
Akers, R.M., McFadden, T.B., Purup, S., Vestergaard, M., Sejrsen, K. and Capuco, A.V., 2000. Local IGF-I axis in peripubertal ruminant mammary development. J. Mammary Gland Biol. Neoplasia, 5: 43–51. https://doi.org/10.1023/A:1009563115612
Akis, I., Oztabak, K., Gonulalp, I., Mengi, A. and Un, C., 2010. IGF-1 and IGF-1R gene polymorphisms in East Anatolian red and South Anatolian red cattle breeds. Russ. J. Genet., 46: 439–442. https://doi.org/10.1134/S1022795410040083
Bartke, A., List, E.O. and Kopchick, J.J., 2016. The somatotropic axis and aging: benefits of endocrine defects. Growth Horm. IGF Res., 27: 41–45. https://doi.org/10.1016/j.ghir.2016.02.002
Bishop, M.D., Tavakkol, A., Threadgill, D.W., Simmen, F.A., Simmen, R.C., Davis, M.E. and Womack, J.E., 1991. Somatic cell mapping and restriction fragment length polymorphism analysis of bovine insulin-like growth factor I. J. Anim. Sci., 69: 4306–4311. https://doi.org/10.2527/1991.69114306x
Bonakdar, E., Rahmani, H.R., Edriss, M.A. and Tabatabaei, S.B., 2010. IGF-I gene polymorphism, but not its blood concentration, is associated with milk fat and protein in Holstein dairy cows. Genet. mol. Res., 9: 1726–1734. https://doi.org/10.4238/vol9-3gmr874
Chung, H.Y., Choi, Y.J., Park, H.N., and Davis, M.E., 2015. Associations between genetic variants in the promoter region of the insulin-like growth factor-1 (IGF1) gene and blood serum IGF1 concentration in Hanwoo cattle. Genet. Mol. Res., 14: 3026-3035. http://dx.doi.org/10.4238/2015.April.10.13
Connor, E.E., Meyer, M.J., Li, R.W., Van Amburgh, M.E., Boisclair, Y.R. and Capuco, A.V., 2007. Regulation of gene expression in the bovine mammary gland by ovarian steroids. J. Dairy Sci., 90: E55–E65. https://doi.org/10.3168/jds.2006-466
Curi, R.A., De Oliveira, H.N., Silveira, A.C. and Lopes, C.R., 2005. Effects of polymorphic microsatellites in the regulatory region of IGF1 and GHR on growth and carcass traits in beef cattle. Anim. Genet., 36: 58–62. https://doi.org/10.1111/j.1365-2052.2004.01226.x
Dai, Z., Wu, F., Yeung, E.W. and Li, Y., 2010. IGF-IEc expression, regulation and biological function in different tissues. Growth Horm. IGF Res., 20: 275–281. https://doi.org/10.1016/j.ghir.2010.03.005
De la Rosa Reyna, X.F., Montoya, H.M., Castrellón, V.V., Rincón, A.M.S., Bracamonte, M.P. and Vera, W.A., 2010. Polymorphisms in the IGF1 gene and their effect on growth traits in Mexican beef cattle. Genet. mol. Res., 9: 875–883. https://doi.org/10.4238/vol9-2gmr745
Ge, W., Davis, M.E. and Hines, H.C., 1997. Two SSCP alleles detected in the 5’-flanking region of bovine IGF1 gene. Anim. Genet., 28: 155–156.
Ge, W., Davis, M.E., Hines, H.C., Irvin, K.M. and Simmen, R.C., 2001. Association of a genetic marker with blood serum insulin-like growth factor-I concentration and growth traits in Angus cattle. J. Anim Sci., 79: 1757–1762. https://doi.org/10.2527/2001.7971757x
Gronostajski, R.M., 2000. Roles of the NFI/CTF gene family in transcription and development. Gene, 249: 31–45. https://doi.org/10.1016/S0378-1119(00)00140-2
Hellström, A., Ley, D., Hansen-Pupp, I., Hallberg, B., Löfqvist, C., van Marter, L., van Weissenbruch, M., Ramenghi, L.A., Beardsall, K. and Dunger, D., 2016. Insulin-like growth factor 1 has multisystem effects on foetal and preterm infant development. Acta Paediatr., 105: 576–586. https://doi.org/10.1111/apa.13350
Islam, K.K., Vinsky, M., Crews, R.E., Okine, E., Moore, S.S., Crews, D.H. and Li, C., 2009. Association analyses of a SNP in the promoter of IGF1 with fat deposition and carcass merit traits in hybrid, Angus and Charolais beef cattle. Anim. Genet., 40: 766–769. https://doi.org/10.1111/j.1365-2052.2009.01912.x
Maj, A., Snochowski, M., Siadkowska, E., Rowinska, B., Lisowski, P., Robakowska-Hyzorek, D., Oprządek, J., Grochowska, R., Kochman, K. and Zwierzchowski, L., 2008. Polymorphism in genes of growth hormone receptor (GHR) and insulin-like growth factor-1 (IGF1) and its association with both the IGF1 expression in liver and its level in blood in Polish Holstein-Friesian cattle. Neuro. Endocrinol. Lett., 29: 981–989.
Miura, S., Tsunoda, N., Ikeda, S., Kai, Y., Cooke, D.W., Lane, M.D. and Ezaki, O., 2004. Nuclear factor 1 regulates adipose tissue-specific expression in the mouse GLUT4 gene. Biochem. biophys. Res. Commun., 325: 812–818. https://doi.org/10.1016/j.bbrc.2004.10.117
Mullen, M.P., Lynch, C.O., Waters, S.M., Howard, D.J., O’boyle, P., Kenny, D.A., Buckley, F., Horan, B. and Diskin, M.G., 2011. Single nucleotide polymorphisms in the growth hormone and insulin-like growth factor-1 genes are associated with milk production, body condition score and fertility traits in dairy cows. Genet. mol. Res., 10: 1819–1830. https://doi.org/10.4238/vol10-3gmr1173
Murney, R., Stelwagen, K., Wheeler, T.T., Margerison, J.K. and Singh, K., 2015. The effects of milking frequency on insulin-like growth factor I signaling within the mammary gland of dairy cows. J. Dairy Sci., 98: 5422–5428. https://doi.org/10.3168/jds.2015-9425
Nagata, K., Guggenheimer, R.A. and Hurwitz, J., 1983. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. natl. Acad. Sci., 80: 6177–6181. https://doi.org/10.1073/pnas.80.20.6177
Nicolini, P., Carriquiry, M. and Meikle, A., 2013. A polymorphism in the insulin-like growth factor 1 gene is associated with postpartum resumption of ovarian cyclicity in Holstein-Friesian cows under grazing conditions. Acta Vet. Scand., 55: 11. https://doi.org/10.1186/1751-0147-55-11
Nowock, J., Borgmeyer, U., Pūschel, A.W., Rupp, R.A. and Sippel, A.E., 1985. The TGGCA protein binds to the MMTV-LTR, the adenovirus origin of replication, and the BK virus enhancer. Nucl. Acids. Res., 13: 2045–2061. https://doi.org/10.1093/nar/13.6.2045
Plath-Gabler, A., Gabler, C., Sinowatz, F., Berisha, B. and Schams, D., 2001. The expression of the IGF family and GH receptor in the bovine mammary gland. J. Endocrinol., 168: 39–48. https://doi.org/10.1677/joe.0.1680039
R Core Team, 2015. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
Ruprechter, G., Carriquiry, M., Ramos, J.M., Pereira, I. and Ana, M., 2011. Metabolic and endocrine profiles and reproductive parameters in dairy cows under grazing conditions: effect of polymorphisms in somatotropic axis genes. Acta Vet. Scand., 53: 35–44. https://doi.org/10.1186/1751-0147-53-35
Siadkowska, E., Zwierzchowski, L., Oprzadek, J., Strzalkowska, N., Bagnicka, E. and Krzyzewski, J., 2006. Effect of polymorphism in IGF-1 gene on production traits in Polish Holstein-Friesian cattle. Anim. Sci. Pap. Rep., 24: 225–237.
Sinnwell, J.P., Therneau, T.M. and Schaid, D.J., 2014. The kinship2 R package for pedigree data. Hum. Hered., 78: 91–93. https://doi.org/10.1159/000363105
Smaragdov, M.G., Prinzenberg, E.M. and Zwierzchowski, L., 2006. QTL mapping in cattle: theoretical and empirical approach. Anim. Sci. Pap. Rep., 24: 69–110.
Szewczuk, M., Zych, S., Czerniawska-Piątkowska, E. and Wójcik, J., 2012. Association between IGF1R/i16/TaqI and IGF1/SnaBI polymorphisms and milk production traits in Polish Holstein-Friesian cows. Anim. Sci. Pap. Rep., 30: 13–24.
Therneaut, M., 2018. Coxme: Mixed effects cox models. R Package Ver., 2: 2-7. https://CRAN.R-project.org/package=coxme.
Wang, Y., Price, S.E. and Jiang, H., 2003. Cloning and characterization of the bovine class 1 and class 2 insulin-like growth factor-I mRNAs. Domest. Anim. Endocrinol., 25: 315–328. https://doi.org/10.1016/j.domaniend.2003.06.001
To share on other social networks, click on any share button. What are these?









