Molecular Cloning, Characterization and Expression Analysis of Wap65 Gene from the Crucian Carp
Molecular Cloning, Characterization and Expression Analysis of Wap65 Gene from the Crucian Carp
Jun Cui, Xiaoxu Zhou, Zhicheng Wang, Derong Kong, Xuemei Qiu, Hongdi Wang and Xiuli Wang*
Nucleotide sequence and deduced amino acid sequence of crucian carpWap65. Nucleotide sequences were determined as described in experimental procedures. The nucleotide sequences are shown below the deduced amino acid sequences. The nucleotide and amino acid numbers are indicated to the left of the sequence. The signal peptide is indicated in the line. The hemopexin-like repeats are indicated by shadows.
Multiple alignment of deduced amino acid sequences for Wap65-like proteins. Abbreviation: Jf, Japanese flounder; Tu, Turbot; Fu, Fugu; Cwl, Chinese weather loach; Chc, Channel catfish; Go, Goldfish; Cc, Crucian carp; Coc, Common carp; HsHPX, Human hemopexin. The other 13Wap65 protein sequences aligned were: Japanese flounder Wap65_1 (Paralichthys olivaceus, KC521544.1), Japanese flounder Wap65_2 (P. olivaceus, KC521545.1), Chinese weather loach Wap65_1 (Misgurnus mizolepis, JN230714.1), Chinese weather loach Wap65_2 (M. mizolepis, JN230715.1), Channel catfish Wap65_1 (Ictalurus punctatus, EU030383.1), Channel catfish Wap65_2 (I. punctatus, EU030384.1), Turbot Wap65_1 (Scophthalmus maximus, KJ160506.1), Turbot Wap65_2 (S. maximus, KJ160507.1), Fugu Wap65_1 (Takifugu rubripes, AB125932.1), Fugu Wap65_2 (T. rubripes, AB125933.1), Common carp Wap65 (Cyprinus carpio, AB052623.1), Goldfish Wap65 (Carassius auratus, D50437.1), Human hemopexin (Genbank ID: NM_000613.2).
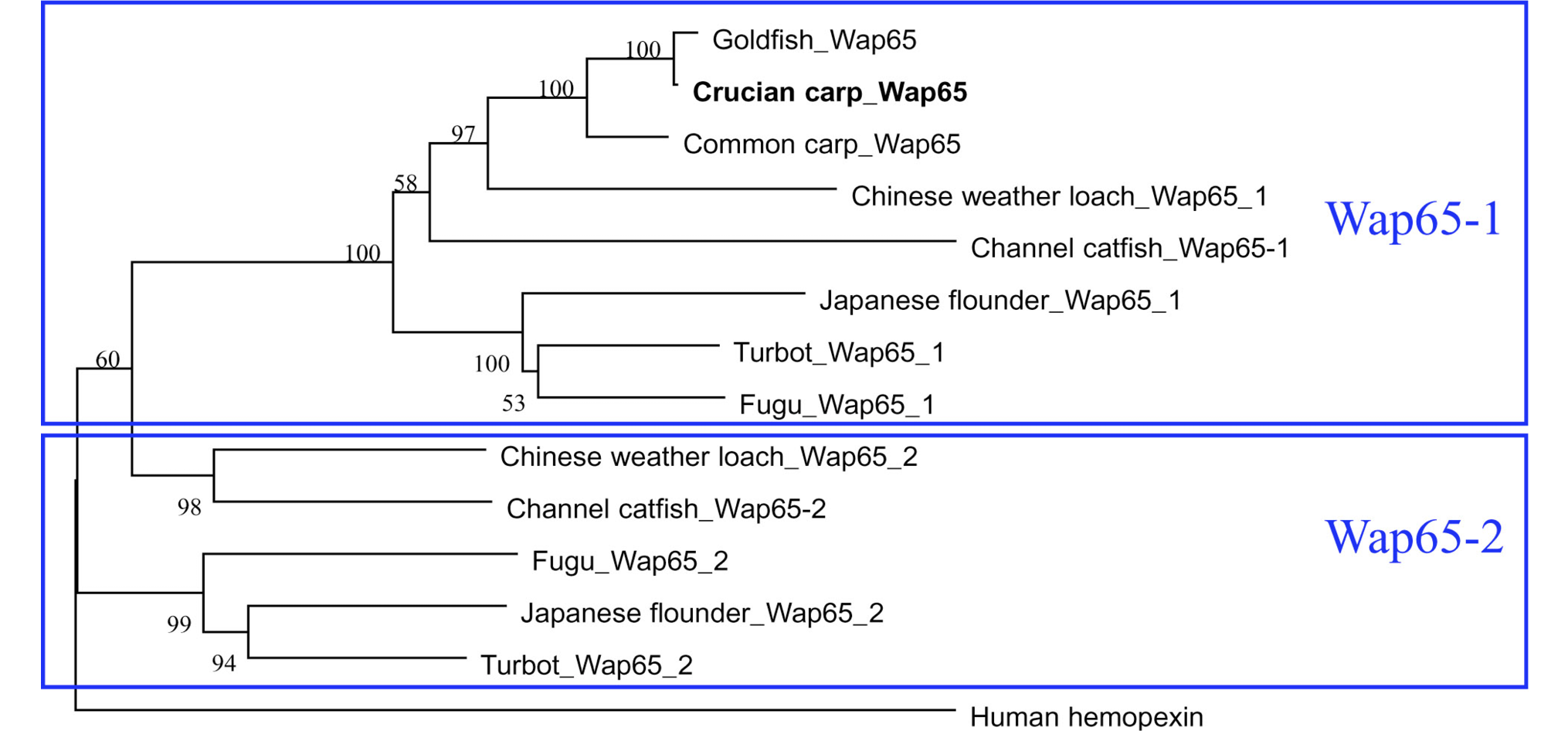
Phylogenetic analysis of Wap65 proteins, including crucian carp Wap65 and another 13 Wap65 protein sequences (Bootstraping=1,000). Neighbor-joining phylogenetic tree was constructed based on the sequence as below utilizing the sequence analysis tool MEGA6, and bootstrapping was performed 1000 times to obtain support values for each branch. The Wap65s were classified into major branches, Wap65-1, Wap65-2 and Human hemopexin.
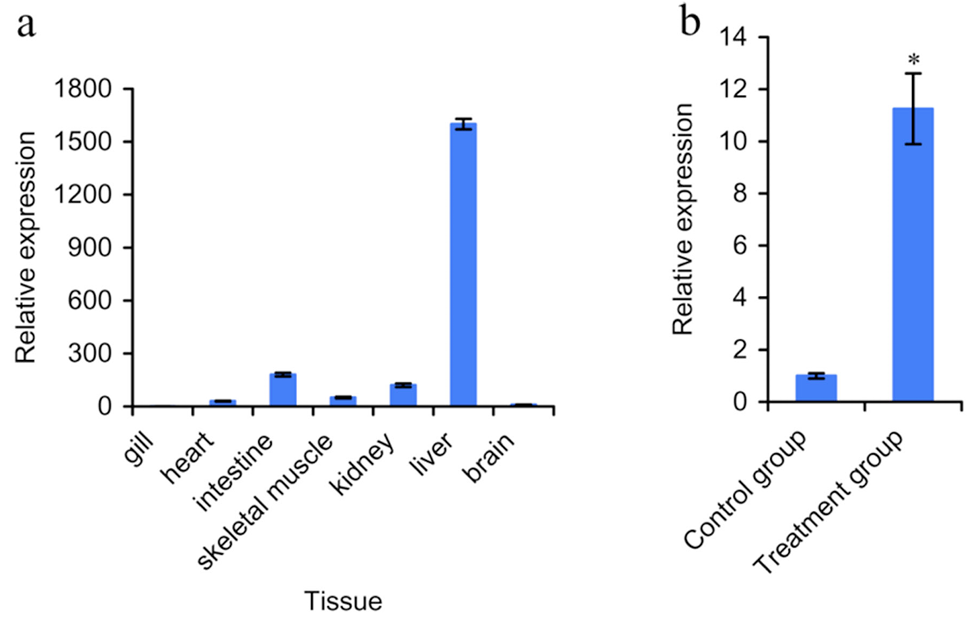
Expression pattern of crucian carp Wap65 mRNA in different experiment. a, Relative expression of Wap65 in crucian carp determined by qRTPCR. The Y-axis represents normalized relative expression values of Wap65. Tissue RNA samples are labeled along the X-axis; b, Fold induction of crucian carp Wap65 gene after heat treatment in the liver. Relative Wap65 expression was expressed as fold change over control samples taken at the same time interval as normalized to change in expression in the β-action control (n=3, *P<0.05).