Kinetic and Parametric Optimization for the Enhanced Production of a Novel Fungal Exo-inulinase under Liquid Culture
Kinetic and Parametric Optimization for the Enhanced Production of a Novel Fungal Exo-inulinase under Liquid Culture
Sikander Ali* and S. Wajiha Khalid
Institute of Industrial Biotechnology, GC University, Lahore
ABSTRACT
The present work describes the optimization of process parameters for the enhanced production of a novel fungal exo-inulinase under liquid culture. Different fungal cultures were isolated and collected from various localities of Lahore and Murree (Punjab, Pakistan). Screening of these cultures was carried out in chemically defined medium i.e., 0.5 g/l (NH4)2SO4, 3 g/l KH2PO4, 1.5 g/l NaNO3, 0.01 g/l MgSO4.7H2O and 3 g/l inulin at pH 7. Kinetic parameters for comparing product yield coefficients of different fungal strains were applied to further optimize exo-inulinase production. The best enzyme producing culture was selected for optimization study. For this, the selected Rhizopus oligosporus (strain ROMII) was used for better enzyme production, for which another screening medium i.e., sucrose minerals salt at pH 5.5 was used. Different physical parameters were applied for maximum enzyme production. The enzyme exhibited maximum activity at pH 6 after 72 h of fermentation (158±0.39 U/ml). The partial purification of the enzyme was done with ammonium sulphate (20-80 %) followed by dialysis which resulted in 21 % purification from the crude enzyme extract. The molecular weight of enzyme was found to be 86 kDa, as determined by SDS poly-acrylamide gel electrophoresis.
Article Information
Received 25 February 2018
Revised 01 May 2019
Accepted 24 June 2019
Available online 06 May 2020
Authors’ Contribution
SA conceived the idea, supervised the research work and finalized the article while SWK did the bench work and compiled the first draft.
Key words
Microbial fermentation, Batch-culture optimization, Kinetic study, Exo-inulinase, Fungal cultures, Submerged fermentation.
DOI: https://dx.doi.org/10.17582/journal.pjz/20180225180228
* Corresponding author: dr.sikanderali@gcu.edu.pk
0030-9923/2020/0005-1657 $ 9.00/0
Copyright 2020 Zoological Society of Pakistan
INTRODUCTION
Inulin in the form of reserved carbohydrate is present in various plants e.g., dahlia, chicory, artichoke and yacon especially in their roots and tubers (Vandamme and Derycke, 1983). It is present in the dried plant tubers in large amounts (Abdulameer et al., 2015). D-fructofuranose with β-2,1-linkages are the linear molecular chains present in inulin that are ended by a glucose unit at the reducing end, to form a sucrose-type linkage (Chi et al., 2009). Inulin hydrolysis mainly yields fructose with little amount of glucose residues. The plants that mostly synthesize and reserve inulin, cannot store starch (Vijayaraghavan et al., 2009). Inulin has significant importance as it is comparatively cheap and ample source for the preparation of fructose syrup, inulooligosaccharide production as well as ethanol fermentation (Naggar et al., 2014). Inulinases (E.C. 3.2.1.7) are the hydrolases that catalyze inulin into glucose and fructose by targeting β-2,1 linkage (Sheng et al., 2007). Two types of inulinases act on inulin, endo-inulinases (2,1-β-D-fructanfructanohydrolase) and exo-inulinases (β-D-fructanfructohydrolase). The endo-inulinase produces inulooligosaccharides as the main product, while exo-inulinase yields fructose as the main product by hydrolyzing the terminal linkages (Kim et al., 2008).
Variety of microbial genera show the ability of producing exo-inulinases, such as Arthrobacter, Bacillus, Clostridium, Pseudomonas, Staphylococcus, Xanthomonas, Aspergillus, Cryptococcus, Penicillium, Sporotrichum, Candida, Kluyveromyces and Pichia (Gill et al., 2003; Bernardo et al., 2008). In general, filamentous fungi show a great potential of producing exo-inulinases because these have been reported to exhibit high enzyme activity in the liquid culture (Mazutti et al., 2010; Adriana et al., 2012). Therefore, the aim of the present work was to isolate and identify filamentous fungal cultures that have the ability to produce extracellular inulinases. The enzyme production was further enhanced by optimization of some kinetic and physical fermentation process parameters.
MATERIALS AND METHODS
The chemicals used in the present study were of analytical grade and procured directly from Fluka (UK) and Sigma (USA). Reagents and other solutions were also of the highest possible purity.
Collection and processing of soil samples
Soil samples from five different localities of Lahore and Murree (Punjab, Pakistan) were collected randomly for isolation of filamentous fungal cultures. Sterile spatula, gloves and plastic bags were used to collect soil samples from a depth of 10-15 cm and brought to IIB immediately. It was ensured that the samples reach to the labs within 24 h of collection. The samples were coded to as B1, B2, B3, B4, and B5. Soil processing was initiated by removing leaves, plant materials, large debris, fibers or other particles from it. Later the samples were stored at 4°C after sealing in sterile polythene bags.
Isolation and screening of filamentous fungal cultures
The filamentous fungal cultures were isolated from their natural habitats by serial dilution method. The medium that was used for isolation contained 0.5 g/l (NH4)2SO4, 3 g/l KH2PO4, 1.5 g/l NaNO3 and 0.01 g/l MgSO4.7H2O at pH 7. However, 3 g/l inulin was sterilized separately and added to the production medium aseptically just before inoculation. The agar plates of the medium were prepared separately by adding 20 g/l agar. The plates were poured and allowed to solidify under aseptic conditions at room temperature. The dilutions of soil samples were made from 10-1 to 10-5 and 1 ml from each of these dilutions was transferred on agar plates. The samples were spread uniformly by rotating the plates clockwise and counter clockwise. The plates were kept at 30°C for 3-5 days in an incubator. The selected strains, with the ability to produce exo-inulinase, were observed under a light microscope for characterization after sub culturing. The slants of fungal cultures were prepared using 40 g/l potato dextrose agar (PDA) medium, pH 5.6.
Characterization and maintenance of fungal isolates
Twelve fungal isolates were selected and maintained for exo-inulinase production. Their optimal growth was accomplished on PDA slants and stored at 4°C. The cultures included Rhizopus oligosporus (ROLI), Aspergillus oryzae (AOLI), A. niger (ANLI), A. flavus (AFLI), Trichoderma sp. (TRLI), Acreonium sp. (ACRLI) and Penicillium sp. (PENLI) from the soils of Lahore, whereas, R. oligosporus (ROMII), A. oryzae (AOMII), A. niger (ANMII), A. flavus (AFMII) and Trichoderma sp. (TRMII) from the soils of Murree.
Inoculum preparation
Spore suspension for each fungal culture was prepared under sterilized conditions. Approximately, 10 ml of autoclaved distilled water was added to a 3-5 d slant culture with profuse growth. The spores were disrupted with a sterilized loop. Later, the tube was gently shaken to form a homogeneous suspension. The spore count was made on a hemocytometer slide bridge, and was found to be 1.2×107 CFU/ml.
Fermentation for exo-inulinase production
Submerged fermentation was used for exo-inulinase production in 250 ml conical flasks. Fifty milliliter of the sucrose mineral salt (SMS) medium containing 15 g/l sucrose, 5 g/l peptone, 1 g/l inulin, 2.5 g/l yeast extract, 0.35 g/l KH2PO4, 0.015 g/l EDTA, 0.0045 g/l CaCl2.2H2O, 0.03 g/l FeSO4.7H2O, 0.003 g/l NaMoO4.2H2O and 0.001 g/l H3BO3 (pH 5.5) was used. All the flasks containing medium were sterilized at 121°C (15 lbs/in2) for 15 min. It was cooled at room temperature and 1 ml of the spore suspension was seeded as an inoculum, under aseptic conditions. Fermentation runs were performed at 30°C with initial pH of 5.5 and 160 rpm in a shaking incubator (VS-8480 Vision, Tokyo, Japan) for 72 h. After termination of run, the flask cultures were taken out from the orbital shaker and the fermented broth was filtered through a pre-weighed Whatmann filter paper (No.1) to remove cellular debris and then dried at 100°C for 30 min (Treichel, 2009). These crude enzyme extracts were centrifuged (PS-EBA-20-E RevA01, Beverly, Massachusetts, USA) at 3500×g for 15 min (4°C), and later used for analysis.
Exo-inulinase assay
One unit of exo-inulinase activity is defined as the amount of enzyme required to release 1 µmol of fructose per milliliter of substrate per minute under standard assay conditions (Cazetta et al., 2005). The crude enzyme extract (0.5 ml) was taken alongwith 2 ml of 0.02 g/l inulin solution and 2 ml of acetate buffer, pH 5.5. The resulting mixture was incubated at 50°C for 20 min. The enzyme reaction was quenched by boiling the reaction mixture in a water bath (1011B, Memmert, Berlin, Germany) for 10 min. Afterwards, the mixture was cooled at room temperature. Two millilitres of 3,5-dinitrosalicylic acid (DNS) reagent was added. It was boiled for another 15 min and later cooled at room temperature. The final volume was raised up to 10 ml by adding 3.5 ml of distilled water. Controls were run in parallel with samples, by replacing crude enzyme extract with the same amount of distilled water. A575 was measured using a spectrophotometer (UV 5100B, China). The enzyme activity was estimated according to Miller (1959) using the following relationship:

Where, DF means the dilution factor while NF means the normalization factor. Protein content was estimated using Bradford (1976) method.
Dry cell mass determination (DCM)
The fermented broth was filtered in a pre-weighed Whatmann filter paper. It was washed twice with distilled water. Later, it was dried in an oven at 70°C for about 20 min. After drying, DCM was calculated by the following relationship,
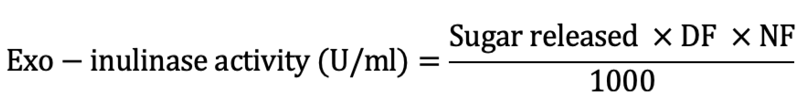
Product yield coefficients
The product yield coefficients i.e., Yp/s (g/g cells) and Yp/x (g/g cells) were determined using the relations, Yp/s (g/g) = dP/ds and Yp/x(g/g) = dP/dX, respectively.
Time duration
Optimal time duration for two fungal cultures i.e., R. oligosporus (ROMII) and Penicillium sp. were compared by incubating the isolates for different time intervals i.e., 12-96 h.
Physical parameters
The effect of various pH (4-7) of SMS medium was studied. The effect of various rate of agitation (40-240 rpm) was studied for maximum enzyme production. The optimal inoculum age was determined by growing fungal cultures for different time periods (24-144 h). Various inoculum sizes (1-6 %) were also compared for maximum enzyme production.
Ammonium sulphate precipitation of exo-inulinase
Exo-Inulinase producing R. oligosporus was grown under the optimal fermentation conditions. The culture broth was harvested after centrifugation (4500×g for 15 min) at 4°C and used for precipitation. It was precipitated by adding 20 % ammonium sulphate and left to stand at 4°C for around 4 h. Afterwards, the extract was centrifuged at 9000×g for 20 min at 4°C. The supernatant obtained was further precipitated using 40, 60 and 80 % ammonium sulphate saturation. The pellets obtained were suspended in 0.01 M acetate buffer at 2:1 (v/w) ratio. This mixture was dialyzed at 4°C overnight with constant stirring until excess ammonium sulphate was removed. The partially purified enzyme was collected.
Molecular weight determination by SDS-PAGE
The molecular weight of exo-inulinase was determined through SDS-PAGE. The sample was diluted with 5X loading dye at the ratio of 1:4 (v/v) and heated at 95oC for 5 min in a water bath. The loading dye gave the sample’s color as blue. Sample was loaded on 12% polyacrylamide gel. Initially the constant voltage of 60 V for 45 min was given. After that, 100 V was provided until the dye touched the base of the resolving gel. The gel was stained in 5% (w/v) coomassie brilliant blue (R-250) for 15 min with slow but continuous stirring and de-stained in 40% (v/v) methanol and 10% (v/v) acetic acid.
RESULTS AND DISCUSSION
Isolation and screening of filamentous fungal strains for exo-inulinase production
A total of 12 filamentous fungal strains were microscopically characterized and identified as shown in Table I. All the strains were grown on a primary screening medium for comparison. The cultures showed clear hollow zones on the culture plates that depicted their ability to produce exo-inulinase. Afterwards, these cultures were grown on a secondary screening medium i.e., sucrose minerals salt (SMS), for fermentation. With each filtrate, dry cell mass, enzyme assay at 575 nm, protein estimation at 595 nm and sugar concentration at 595 nm were determined. Among the kinetic parameters the product yield coefficients (Yp/x and Yp/s) were also determined. Different cultures exhibited a range of diverse physical appearance of colonies. The results depicted that the best enzyme producing strain was ROMII with the enzyme activity i.e., 27±0.06 U/ml which was 6.75-fold higher as compared to the strain ACRLI (4±0.01 U/ml). The other better strain was PENLI with 21±0.05 U/ml of enzyme activity having 5.25-fold higher activity as compared to the strain ACRLI. Similarly, the values of protein content, dry cell mass, sugar concentration, Yp/x and Yp/s from the strain ROMII were higher as compared to other strains (42±0.10 mg/ml, 24±0.06 g/l, 36±0.09 mg/ml, 1.54±0.07 and 0.95±0.04, respectively). However, for strain PENLI the values were found to be 38±0.09 mg/ml, 21±0.05 g/l, 31±0.07 mg/ml, 1.42±0.07 and 0.87±0.04, respectively as shown in Figure 1. The cell count of strain PENLI was found to be 2.9×105 CFU/ml and for that of strain ROMII was 5.9×105 CFU/ml, that was over 2 fold higher than that of strain PENLI and 4.5 fold higher than strain ACRLI having 1.3×105 CFU/ml. Ohta et al. (2002) and Mohamed et al. (2015) reported R. oligosporus to be the best enzyme producing microorganism in their respective findings.
Time of incubation
The optimal time duration of the two selected cultures ROMII and PENLI was determined and compared. Both strains showed active exo-inulinase production in 72 h. The highest enzyme activity for strains ROMII and PENLI at 72 h was obtained as 30±0.07 and 22±0.05 U/ml, respectively (Fig. 2). The fold of enzyme production of strain ROMII was 7.5 times higher than that of strain PENLI due to which it was selected for optimization studies. The value for protein content of strain ROMII at 72 h i.e., 44±0.11 mg/ml, was 1.22 fold higher than that of strain PENLI (36±0.09 mg/ml). The sugar consumption
Table I.- Isolation and screening of best filamentous fungal culture for the production of inulinases.
|
Sr. No. |
Culture isolates |
Codes |
Physical appearance of colonies |
Hemocytometer calculation (CFU/ml) |
Inulinase activity (U/ml) |
Yp/x (g/g cells) |
Yp/s (g/g) |
|
Lahore |
|||||||
|
1 |
Rhizopus oligosporus |
ROLI |
White circular |
5.1×105 |
18±0.04 |
0.9±0.04 |
0.64±0.03 |
|
2 |
Aspergillus oryzae |
AOLI |
Dumpy greenish |
4.4×105 |
17±0.04 |
1.1±0.05 |
0.68±0.03 |
|
3 |
Aspergillus niger |
ANLI |
Dumpy greenish |
3.7×105 |
15±0.03 |
1.2±0.06 |
0.68±0.03 |
|
4 |
Aspergillus flavus |
AFLI |
Dumpy greenish |
2.8×105 |
12±0.03 |
1.2±0.06 |
0.66±0.03 |
|
5 |
Trichoderma spp. |
TRLI |
Dumpy greenish |
2.3×105 |
9±0.02 |
1.3±0.06 |
0.69±0.02 |
|
6 |
Acreonium spp. |
ACRLI |
Dumpy grayish |
1.3×105 |
4±0.01 |
1.1±0.06 |
0.4±0.04 |
|
7 |
Penicillium spp. |
PENLI |
Yellow mass |
2.9×105 |
21±0.05 |
1.4±0.07 |
0.87±0.04 |
|
Murree |
|||||||
|
8 |
Rhizopus oligosporus |
ROMII |
Yellow circular mass |
5.9×105 |
27±0.06 |
1.5±0.07 |
0.95±0.04 |
|
9 |
Aspergillus oryzae |
AOMII |
Greenish small clusters |
4.1×105 |
16±0.04 |
1.1±0.05 |
0.84±0.04 |
|
10 |
Aspergillus niger |
ANMII |
Greyish small clusters |
3.4×105 |
13±0.03 |
1.3±0.06 |
0.82±0.04 |
|
11 |
Aspergillus flavus |
AFMII |
Greenish and whitish small clusters |
2.1×105 |
10±0.02 |
1.3±0.06 |
0.81±0.04 |
|
12 |
Trichoderma spp. |
TRMII |
White small clusters |
2.2×105 |
6±0.01 |
1.3±0.06 |
0.66±0.03 |
Incubation temperature 30°C, pH 5.5, time 72 h, agitation 160 rpm, inulin conc. 1 g/l, inoculum age 1 d, inoculum size 2 %. Yp/x (g/g cells), dP/dX; Yp/s (g/g), dP/ds. Each value is a mean average of three parallel replicates. ± indicate the 5 % standard deviation.
in 72 h by strain ROMII (39±0.09 mg/ml) was 1.18 fold higher as compared to strain PENLI at 72 h (33±0.08 mg/ml). Similarly, DCM obtained for strain ROMII in 72 h (28±0.07 g/l), was 1.16 fold higher than that of strain PENLI (24±0.05 g/l). But for both the cultures, the value of DCM increased with the increase in time and it was due to prolonged fermentation. Kumar et al. (2005) also reported maximum enzyme production at 72 h of incubation from A. niger.
Effect of initial pH
The maximum enzyme activity attained at pH 6 was 83±0.2 U/ml with protein content (88±0.22 mg/ml) by strain ROMII. The sugar concentration and DCM values at this pH were obtained as 58±0.14 mg/ml and 36±0.09 g/l, respectively. Gill et al. (2006) also reported the maximum exo-inulinase activity at pH 6 from A. fumigatus. The enzyme activity was about 1.76 fold higher as compared to the activity at other pH values, and the protein content was almost 1.8 fold higher, as shown in the Figure 3. The change in pH had an adverse effect on the enzyme activity, a very low activity was observed at lower pH values which showed that the enzyme was highly sensitive to pH changes. Whereas, the enzyme activity was slightly affected at pH ranges near to neutral. Similarly, the values for protein content, sugar concentration and DCM also had a similar effect to pH changes, being low at lower pH values.
Effect of agitation rate
Maximum exo-inulinase activity was obtained at 120 rpm i.e., 93±0.23 U/ml by strain ROMII. The protein content at this agitation rate was also higher i.e., 98±0.24 mg/ml. The values for sugar concentration and DCM were 65±0.16 mg/ml and 39±0.09 g/l, respectively. The enzyme activity and protein content were 1.55 and 1.45 fold higher as compared to values at other agitation rates, as shown in the Figure 4. Almost insignificant enzyme activity was exhibited at 40 rpm i.e., 64±0.16 U/ml. Similarly, the values for enzyme activity, protein content, sugar concentration and DCM, at higher agitation rates were not adversely affected as compared to the values at low agitation rate. Gallegos et al. (2014) reported 200 rpm for maximum enzyme activity from R. microspores.
Effect of size and age of inoculum
The maximum exo-inulinase activity was exhibited by the strain ROMII that was incubated for 2 d i.e., 102±0.25 U/ml. The protein content, sugar concentration and DCM were also higher at this inoculum age i.e., 112±0.28 mg/ml, 71±0.17 mg/ml and 41±0.10 g/l, respectively. The enzyme activity from a 2 d incubated culture was 1.07 fold higher than that shown by 1 d as shown in Figure 5. Onulide et al. (2012) reported maximum enzyme activity from a 24 h old culture of Saccharomyces sp. The best inoculum size of ROMII was determined to be 5% as it supported maximum enzyme activity (119±0.29 U/ml). This value was almost 1.2 fold higher (101±0.23 U/ml) as compared to the value of 2% inoculum level previously being used. The values for protein content, sugar concentration and DCM at 5% inoculum size were, 128±0.32 mg/ml, 83±0.21 mg/ml and 54±0.13 g/l, respectively. These values were 1.2, 1.48 and 2.45 fold higher as compared to values at other inoculum sizes. Kumar et al. (2005) reported better enzyme production at 10% inoculum by A. niger.
Ammonium sulphate precipitation
The maximum exo-inulinase activity was obtained at 80% ammonium sulphate saturation i.e., 152±0.44 U/ml. It was 1.23 fold higher than the value obtained at 40% ammonium sulphate saturation. Similarly, the protein content (159±0.39 mg/ml) at 80% ammonium sulphate concentration was 1.11 fold higher than at 40% precipitation. The specific activity was also higher at 80% saturation (0.95 U/mg), as depicted in Table II. The specific activity at 80% saturation was 1.16 fold higher as compared to that at 40% saturation level. The values gradually decreased after dialysis, however the value of specific activity of 80% saturation increased after dialysis i.e., 0.97 U/mg. The fold-purification after dialysis was maximum i.e., 0.99 fold whereas the yield obtained for the enzyme activity was recorded to be 21%. El-souod et al. (2014) reported partial purification of an exo-inulinase through 80 % ammonium sulphate precipitation.
Determination of molecular weight of enzyme
The molecular weight of partially purified exo-inulinase from strain ROMII was estimated with SDS-PAGE. A single protein band of molecular weight 86 kDa was obtained as shown in Fig. 6. This indicated an oligomeric form of the enzyme. The molecular weight of an exo-inulinase from R. oligosporus TN-96 by Ohta et al. (2002) was 83 kDa and that of R. oligosporus NRRL 2710 was 76 kDa as reported by Mohamed et al. (2015). Yedahalli et al. (2016) reported molecular weight to be 81 kDa.
Table II.- Activity of inulinase isolated from R. oligosporus (ROMII) with different concentrations of ammonium sulphate before and after dialysis.
|
Ammonium sulfate conc. (%) |
Before dialysis |
Inulinase activity (U/ml) after dialysis |
||
|
Inulinase activity (U/ml) |
Protein (mg/ml) |
Specific activity (U/mg) |
||
|
20 |
156±0.39 |
165±0.41 |
0.94±0.04 |
140±0.35 |
|
30 |
116±0.29 |
137±0.34 |
0.84±0.04 |
109±0.27 |
|
60 |
108±0.27 |
124±0.31 |
0.87±0.04 |
78±0.19 |
|
80 |
152±0.38 |
159±0.39 |
0.95±0.04 |
129±0.32 |
Incubation temperature 30°C, pH 6, time 72 h, agitation 120 rpm, inulin conc 1 g/l, inoculum age 2 d, inoculum size 5 %. Each value is a mean average of three parallel replicates, ± indicated 5% standard deviation among these values.
CONCLUSIONS
In this study, fungal exo-inulinase was produced substantially. Kinetic studies revealed R. oligosporus (strain ROMII) to be more efficient strain for enzyme production. The maximum enzyme activity (158±0.39 U/ml) was 5.81 fold improved after the optimization of process parameters by ROMII under liquid culture. Later, the enzyme was partially purified and its molecular weight was found to be 86 kDa. These results are highly significant (HS) and exhibit a viable potential after scale up studies.
ACKNOWLEDGEMENTS
We are grateful to the Director IIB and Vice Chancellor of the University for providing research facilities and moral assistance.
Conflict of interest declaration
The authors declare that they have no competing interests.
REFERENCES
Abdulameer, Z.W., Essa, B.N. and Aziz, G.M., 2015. Optimum conditions for inulinase production by Aspergillus niger using solid state fermentation. Baghdad Sci. J., 12: 1-10. https://doi.org/10.21123/bsj.12.2.307-316
Adriana, C., Flores, G., Jesus, M.C., Cristobal, N.A. and Raul, R.H., 2012. Inulinase production by a Mexican semi desert xerophilic Penicillium citrinum strain under submerged culture. J. Fd. Sci. Technol., 4: 46-50.
Bernardo, O., Silva-Santisteban, Y., Converti, A. and Filho, F.M., 2008. Effects of carbon and nitrogen sources and oxygenation on the production of inulinase by Kluyveromyces marxianus. Appl. Biochem. Biotechnol., 152: 249-261. https://doi.org/10.1007/s12010-008-8247-7
Bradford, M.M., 1976. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72: 248-254.
Cazetta, M.L., Martins, P.M.M., Monti, R. and Contiero, J., 2005. Yacon (Polymnia sanchifolia) extract as a substrate to produce inulinase by Kluyveromyces marxianus var. bulgaricus. J. Fd. Eng., 66: 301-305. https://doi.org/10.1016/j.jfoodeng.2004.03.022
Chi, Z., Chi, Z., Zhang, T., Liu, G. and Yue, L., 2009. Inulinase-expressing microorganisms and applications of inulinases. Appl. Microbiol. Biotechnol., 82: 211-220. https://doi.org/10.1007/s00253-008-1827-1
El-Souod, S.M., Mohamed, T.M., Ali, E.M.M., El-Badry, M.O. and El-Keiy, M.M., 2014. Partial purification of extracellular exo-inulinase from Ulocladium atrum. J. Genet. Eng. Biotechnol., 12: 15-20. https://doi.org/10.1016/j.jgeb.2014.04.001
Gallegos, A.C., Contreras-Esquivel, J.C., Morlett-Chavez, J.A., Aguilar, C.N. and Rodriguez-Herrera, R., 2014. Comparative study of fungal strains for thermostable inulinase production. J. Biosci. Bioengin., 1: 1-6.
Gill, P.K., Sharma, A.D., Harchand, R.K. and Singh, P., 2003. Effect of media supplements and culture conditions on inulinase production by an Actinomycete strain. Bioresour. Technol., 87: 359-362. https://doi.org/10.1016/S0960-8524(02)00262-6
Gill, P.K., Manhas, R.K. and Singh, P., 2006. Hydrolysis of inulin by immobilized thermostable extracellular exoinulinase from Aspergillus fumigatus. J. Fd. Eng., 76: 369-375. https://doi.org/10.1016/j.jfoodeng.2005.05.052
Kim, K.Y., Nascimento, A.S., Golubev, A.M., Polikarpov, I., Kim, C.S., Kang, S. and Kim, S., 2008. Catalytic mechanism of inulinase from Arthrobacter sp. S37. Biochem. biophys. Res. Commun., 371: 600-605. https://doi.org/10.1016/j.bbrc.2008.03.126
Kumar, G.P., Kunamneni, A., Prabhakar, T. and Ellaiah, P., 2005. Optimization of process parameters for the production of inulinase from a newly isolated Aspergillus niger AUP19. World J. Microbiol. Biotechnol., 21: 1359-1361. https://doi.org/10.1007/s11274-005-5078-3
Mazutti, M.A., Skrowonski, A., Boni, G., Zabot, G.L., Silva, M.F., Oliveira, D., Luccio, M.D., Filho, F.M., Rodrigues, M.I. and Treichel, H., 2010. Partial characterization of inulinases obtained by submerged and solid-state fermentation using agro industrial residues as substrates: a comparative study. Appl. Biochem. Biotechnol., 160: 682-693. https://doi.org/10.1007/s12010-009-8687-8
Miller, G.L., 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal. Chem., 31: 426-428. https://doi.org/10.1021/ac60147a030
Mohamed, S.A., Salah, H.A., Moharam, M.E., Foda, M.S. and Fahmy, A.S., 2015. Characterization of two thermostable inulinases from Rhizopus oligosporus NRRL 2710. J. Genet. Eng. Biotechnol., 13: 65-69 https://doi.org/10.1016/j.jgeb.2014.12.001.
Naggar, N.A., Metwally, E.A., Tanash, A.B. and Sherief, A.A., 2014. Screening of inulinolytic potentialities of some fungi isolated from Egyptian soil. Biotechnology, 13: 152-158. https://doi.org/10.3923/biotech.2014.152.158
Ohta, K., Hamada, S. and Nakamur, T., 1993. Production of high concentrations of ethanol from inulin by simultaneous saccharification and fermentation using Aspergillus niger and Saccharomyces cerevisiae. Appl. environ. Microbiol., 59: 729-738.
Onulide, A.A., Fadaunsi, I.F. and Garuba, E.O., 2012. Inulinase production by Saccharomyces sp. in solid state fermentation using wheat bran as substrate. Annls. Microbiol., 62: 843-848. https://doi.org/10.1007/s13213-011-0325-3
Sheng, J., Chi, Z.M., Li, J.L.M. and Gong, F.G., 2007. Inulinase production by the marine yeast Cryptococcus aureus G7a and inulin hydrolysis by the crude inulinase. Proc. Biochem., 42: 805-811. https://doi.org/10.1016/j.procbio.2007.01.016
Treichel, H., Mazutti, M.A., Maugeri, F. and Rodrigues, M.I., 2009. Use of a sequential strategy of experimental design to optimize the inulinase production in a batch bioreactor. J. Indus. Microbiol. Biotechnol., 36: 895-900. https://doi.org/10.1007/s10295-009-0567-2
Vandamme, E.J. and Derycke, D.G., 1983. Microbial inulinases: Fermentation process, properties and applications. Adv. appl. Microbiol., 29: 139-176. https://doi.org/10.1016/S0065-2164(08)70356-3
Vijayaraghavan, K., Yamini, D., Ambika, V. and Sowdamini, N.S., 2009. Trends in inulinase production. Cri. Rev. Biotechnol., 29: 67-77. https://doi.org/10.1080/07388550802685389
Yedahalli, S.S., Rehmann, L. and Bassi, A., 2016. Expression of exo-inulinase gene from Aspergillus niger 12 in E. coli strain Rosetta-gami B (DE3) and its characterization. Biocatal. Bioreact. Des., 32: 629-637. https://doi.org/10.1002/btpr.2238
To share on other social networks, click on any share button. What are these?










