Genetic Diversity of Mycobacterium tuberculosis Complex Prevailing In Khyber Pakhtunkhwa, Pakistan
Genetic Diversity of Mycobacterium tuberculosis Complex Prevailing In Khyber Pakhtunkhwa, Pakistan
Bashir Ahmad1, Muhammad Idrees1,*, Kafeel Ahmad1, Dawood Ahmad2, Sajid Ali3 and Shumaila Bashir4
1Centre of Biotechnology and Microbiology, University of Peshawar, Peshawar
2Institute of Biotechnology and Genetic Engineering, The University of Agriculture, Peshawar
3TB Reference Laboratory, Peshawar, Khyber Pakhtunkhwa
4Department of Pharmacy, University of Peshawar, Peshawar
ABSTRACT
The Khyber Pakhtunkhwa region of Pakistan has been known for a high burden of tuberculosis (TB). However, there is little information about the molecular characteristics of Mycobacterium tuberculosis (M. tuberculosis) strains predominant in the province. Therefore, this study was planned to study the genetic diversity in the M. tuberculosis isolates isolated from clinical samples. In the current study total 794 patients were tested for suspected tuberculosis by fluorescence microscopy and GeneXpert system during the study period (2015-2016) at the Provincial TB Reference Laboratory, Khyber Pakhtunkhwa, Pakistan. Among the 794 patients studied, 170 samples gave positive results. Fifty-two Mycobacterium tuberculosis complex (MTBC) isolates were randomly selected for RAPD-PCR and PCR-restriction enzyme analysis. Mycobacterium bovis (M. bovis) and M. tuberculosis H37Rv were used as reference strains. RAPD-PCR differentiated the strains and authenticated the high molecular diversity. PCR-restriction enzyme analysis (PRA) profiles of all isolates and reference strains were found to be the same. PRA is a valid alternative to phenotypic identification in a routine diagnostic laboratory. High diversity values were found in the studied area implying high population diversity of M. tuberculosis that’s being prevalent in Khyber Pakhtunkhwa, Pakistan.
Article Information
Received 14 October 2017
Revised 20 November 2017
Accepted 25 December 2017
Available online 15 March 2018
Authors’ Contribution
KA and BA presented the concept and designed the study. MI, SA and KA performed practical work. DA and KA analyzed the data. KA and MI wrote the manuscript. SB and KA proofread the final draft.
Key words
Tuberculosis, PCR, Diversity, GeneXpert, Microscopy.
DOI: http://dx.doi.org/10.17582/journal.pjz/2018.50.2.663.669
* Corresponding author: m.idrees436@yahoo.com
0030-9923/2018/0002-0663 $ 9.00/0
Copyright 2018 Zoological Society of Pakistan
Introduction
The causative agent of the disastrous tuberculosis disease is Mycobacterium tuberculosis complex (MTBC). MTBC consists of a genetically related group of Mycobacterium species that cause tuberculosis in humans and a range of other organisms. There are around 130 species in the Genus Mycobacterium, most of them are causative agents for the disease in human (Tortoli et al., 2009). Among them are M. africanum, M. cannetii, M. microti, M. bovis and M. tuberculosis. Humans are the natural hosts of M. tuberculosis and M. africanum whereas M. bovis commonly infect domestic and wild animals along with humans. Among the major zoonotic diseases that pose public health threats with excessive socio-economic effect are bovine tuberculosis and brucellosis (Batool et al., 2017). To identify MTBC species correctly is paramount for the distinction between human and zoonotic TB.
In most developing countries microscopy is the only available method for identification of MTBC. Moreover, it is rapid, simple, and economical for the detection of acid fast bacillus (AFB) present in TB patients. It can be also be performed using Ziehl–Neelsen (ZN) staining or fluorescence microscopy (FM) using a fluorescent substance such as auramine phenol to increase the visualization (Ogbaini-Emovon, 2009)
GeneXpert MTB/RIF is a molecular technique for the diagnosis of TB. The technique not only detects M. tuberculosis but also identifies the presence or absence of various drug resistant mutations in DNA fragments like rifampicin resistance (Marlowe et al., 2011).
At present various molecular techniques are used for genotyping of M. tuberculosis such as IS6110-RFLP, MIRU-VNTR, and spoligotyping. All these techniques are quite complex, time-consuming and expensive (Guillén-Nepita et al., 2013; Jonsson et al., 2014). Hence an economical and simple method such as random amplification of polymorphic DNA (RAPD) is sought after analyzing the diversity of the mycobacterial genome in low-income countries (Guillén-Nepita et al., 2013). Currently, RAPD-PCR is being used to characterize the pathogen and elucidate the phylogeny of diverse animal and plant species. The technique has high specificity but experiences certain demerits such as less reproducibility. Still, RAPD-PCR is a relatively simple, easy and valuable tool in epidemiological studies. The hsp65 gene has been reported as a relevant target in the identification of Mycobacterium (Kim et al., 2005). The polymerase chain reaction restriction enzyme analysis (PRA) efficiently discriminates between and identifies Mycobacterium species. The partial hsp65 gene (604bp) is the most preferred choice for the identification and phylogenetic analysis of Mycobacterium species (Kim et al., 2005).
The evaluation of genetic diversity within M. tuberculosis is very critical. Genetic diversity determines variability in disease progression, frequency of transmission, and response to the treatment. An understanding of these factors is imperative to characterize TB and provide the most suitable treatment. The genetic diversity also delineates evolutionary relationship among bacteria.
Tuberculosis cases are more common in countries like Pakistan and there is little information about the molecular characteristics of M. tuberculosis strains predominant in the Khyber Pakhtunkhwa province. Therefore, this study was planned to study the genetic diversity in the M. tuberculosis isolates isolated from clinical samples.
Materials and Methods
Samples collection and processing
Informed consent was taken from all participating patients as approved by ethical committee of the Centre of Biotechnology and Microbiology, University of Peshawar. During the study (2015-2016), a total of 794 sputum samples (462 from male and 332 from female) were collected from various geographic locations of Khyber Pakhtunkhwa at Provincial TB Reference Laboratory (PTRL). The patients involved in the study were of different age groups. Standard procedure was followed for collecting a sputum sample from tuberculosis patients (Varaine et al., 2010). The sputum specimens (2-3 mL) were collected in 50 mL sterile plastic vials. The patients were instructed not to touch the inside of the collection bottle or lid. The sample vials were opened only in a biosafety cabinet. Sputum samples were processed within 24 h of collection. The majority of the patients were recently treated, while some were from the previously treated groups. All of the participating patients were HIV negative. The standard protocol was followed for the digestion and decontamination of samples (Kent and Kubica, 1985; Siddiqi and Rüsch-Gerdes, 2006).
Identification of MTBC by fluorescence microscopy
A standard protocol was followed to prepare slides for each sample (Initiative, 2014). Briefly, a small amount of sample was spread onto slides using a sterile inoculating loop and heat fixed by passing two to three times over flames. Followed by this, heat fixation slides were stained with auramine/rhodamine for 20 min and then slides were rinsed with distilled water. After this, the slides were treated with 0.5% alcohol for 2 min followed by the treatment with 0.5% potassium permanganate for 1−2 min. Finally, the slides were washed; air dried and observed immediately using a fluorescence microscope.
GeneXpert assay
A sputum sample was mixed with decontamination reagent in a 2:1 (v/v) ratio for 15 min at ambient temperature with intermittent mixing. The mixture was then loaded into the cartridge and placed in GeneXpert instrument (GX). Results were ready within two hours. Sensitivity and specificity of the fluorescence microscopy and GeneXpert assay were calculated as follows:
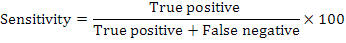
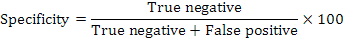
Culturing of MTBC
BACTEC MGIT 960 system (Cat No: 445870) was used for culturing of MTBC. Lowenstein-Jensen (LJ) slants was also used for primary isolation.
Immunochromatographic test
The chromatographic immunoassay was carried out as per manufacturer’s instructions (BD MGIT TBc Identification Test). TBc ID devices (Catalog No: 245159, Becton, Dickinson) were taken out from the foil pouch and were placed on a flat surface. The devices were labeled appropriately as per samples to be tested. The samples were mixed gently by vortexing and 100 µL of sample was loaded on the appropriately labeled device well using a sterile pipette. The results of the test were observed and recorded after 15 min.
Genetic diversity identification (RAPD-PCR)
Fifty-two clinical isolates of mycobacteria from human patients were randomly selected to evaluate the genetic diversity in MTBC. All isolates were cultured and maintained under controlled conditions. The DNA was extracted from the colonies of mycobacteria by combining the method of heat and sonication as mentioned earlier. DNA amplification of mycobacteria was carried out in a DNA amplifier [Eppendorf AG 22331 Hamburg] using random primers as specified in Table I. Thermocycler conditions were programmed as recommended by the manufacturer (Solis BioDyne–5X FIREPol Master mix). Each PCR reaction in 25 µL contained 0.3 μL of primer, 4 μL of master mix, 2 μL of genomic DNA and 18.7 μL of molecular grade water. The amplification conditions were programmed as 5 min initial denaturation step at 94°C, denaturation at 94°C for 1 min, annealing at 36°C for 1 min and elongation at 72°C for 1 min. The number of reaction cycles was set to 45. The final step of elongation at 72°C for 5 min was also incorporated. PCR amplified products were visualized using gel electrophoresis. Standard DNA ladder of 1 kb (Cat No: SM0313) was used for comparison and determination of amplicon size. The gel was observed and photographed using a gel-doc system (SYNGENE Serial number SYDR/2138).
Table I.- Primers used in RAPD-PCR.
| Primer | Sequence | Reference |
| OPN–01 | 5’CTCACGTTGG3’ | |
| OPN–02 | 5’ACCAGGGGCA3’ | |
| OPN–20 | 5’GGTGCTCCGT3’ |
Table II.- Primers used for amplification of hsp65 gene (Telenti et al., 1993).
| Gene | Primer | Sequence |
Product size (bp) |
| hsp65 | Tb11 | ACCAACGATGGTGTGTCCAT |
439 |
| Tb12 | CTTGTCGAACCGCATACCCT |
Statistical analysis
A data matrix, consisting of the number 1 and 0, was assembled on the basis of presence (1) or absence (0) of a DNA band appearing in each lane. The data were used to evaluate of genetic diversity among MTBCs. The genetic divergence among the isolates was determined using the formula Dxy = 1-Nxy/ (Nx+Ny−Nxy) (Nei and Li, 1979). In this formula, Dxy is the score of difference between isolates X and Y. Similarly, Nxy is the number of similar bands between isolates X and Y genotypes. While Nx is the entire number of bands present in the isolate X and Ny is the total number of bands present in the isolate Y. These data were also used for constructing a dendrogram using NTSYS-PC software version 2.10e (Rohlf, 2000).
PCR restriction enzyme analysis (PRA)
RAPD-PCR was performed for the 52 clinical isolates of mycobacteria that were further selected for PRA.
Partial hsp65 gene amplification for PRA
The amplification of the partial hsp65 gene (439 bp) was performed using primers (Table II) as described by Telenti et al (1993). The PCR protocol for the amplification was as follows: PCR reaction mixture (27 μL) consisted of 4 μL of master mix, 0.3 μL of forward and reverse primer each, 5 μL DNA sample and 17.4 μL molecular grade water. PCR thermal program was set as an initial denaturation at 95°C for 1 min, denaturation at 94°C for 1 min, annealing at 60°C for 1 min and elongation at 72°C for 1 min. The number of reaction cycles was set to 44 and the last step of extension at 72°C for 10 min was also included. After completion of the amplification process, the resultant amplified products were analyzed by agarose gel electrophoresis. A standard DNA ladder of 1 kb was used for the comparison and determination of the sizes of amplified products.
Bsu RI treatment
In a fresh sterile tube, the restriction digestion reaction was set by adding 10 µL of PCR amplified product of hsp65 gene, 18 µL of PCR grade water, 2 µL of 10x Buffer R, and 1.5 µL of BsuRI (HaeIII) (#ER0151). The reaction mixture was mixed gently for a few seconds and incubated at 37°C for 2-3 h.
Eco91I treatment
In a fresh sterile tube, the restriction digestion reaction was set by adding 10 µL of PCR amplified products of hsp65 gene, 18 µL of PCR grade water, 2 µL 10x Buffer O and 1.5 µL of Eco91I (BstEII) (#ER0391). The reaction mixture was mixed gently for a few seconds and incubated at 37°C for 2-3h.
2% Agarose Gel Electrophoresis
After restriction digestion with selected enzymes, the resultant fragments were evaluated on a 2% agarose gel. A standard 100 bp of DNA ladder (#SM0323) was used for size comparison. The generated restriction patterns of the isolates were verified against those present in PRA online database (http://app.chuv.ch/prasite/index.html) and identification of Mycobacterium species was performed.
Results
Identification of MTBC
Among the 794 patients investigated by fluorescence microscopy, 111 (13.97%) were observed to be MTBC-positive. Out of the 794 patients examined by GeneXpert assay, 170 (21.4%) were found to be MTBC positive. Among these, 59 cases that were identified as negative on the basis of microscopy were detected as positive by the GeneXpert assay. In comparison with fluorescence microscopy, the sensitivity and specificity of GeneXpert were evaluated. The sensitivity of the fluorescence microscopy was 74.23% while the sensitivity of the GeneXpert was 99.90% . The specificity of both techniques was found as 99.9%. Figure 1 shows the comparative evaluation of the sensitivity of fluorescence microscopy and GeneXpert assay.
Immunochromatographic test
About 170 samples were screened using the immunochromatographic method. Positive immunochromatographic test results appear as shown in Figure 2 . Among 170 samples examined, only 5 samples showed negative results, i.e., at the “T” position no red to the pink line was observed.
RAPD-PCR
All primers used in RAPD-PCR generated polymorphic bands. Different DNA fragments in each of the isolates were amplified and it was found that most of the fragments were common to different strains. Amplicons’ size ranged between 250 and 1200 bp. All 52 isolates showed a high degree of polymorphisms. The level of genetic polymorphism was from 0 to 100%. Table III shows RAPD patterns observed for MTBC isolates.
Table III.- RAPD patterns of MTBC isolates.
| Primer |
No of alleles |
Polymorphic bands |
Percent polymorphism |
Allele size |
| OPN–01 |
5 |
5 |
100% |
250-900 |
| OPN–02 |
6 |
6 |
100% |
250-1200 |
| OPN–20 |
5 |
5 |
100% |
300-750 |
| Total |
16 |
16 |
100% |
250-1200 |
Phylogenetic analysis
Bivariate data analysis was applied to construct dendrograms for all RAPD primers (Fig. 3). Two major clusters, A and B, were formed. Cluster A was composed of only three members: M1, M44, and M5. Being a larger one, Cluster B, was subdivided into three sub clusters: B1, B2 and B3. Group B1 was further divided into two sub clusters: B1a and B1b. Sub cluster B1a was comprised of 24 isolates: M2, M20, M46, M48, M33, M49, M43, M39, M4, M42, M29, M35, M8, M14, M45, M17, M24, M19, M18, M28, M7, M47, M10, and M37. Sub cluster B1b contained 17 isolates: M9, M11, M13, M16, M50, M25, M23, M38, M12, M15, M21, M30, M40, M22, M36, M26, and M27. While, B2 cluster was composed of only three members: M3, M34, and M52. Cluster B3 consists of only two members: M31 and M32. However, three strains, viz., M51, M41, and M6, were not placed in any cluster and considered as operational taxonomy units (OTUs). It can be concluded that RAPD markers are competent tools to estimate genetic diversity and relationships among MTBC.
Amplification of hsp65 gene
Partial hsp65 gene (439bp) was observed to be amplified in 52 clinical isolates. Figure 4 shows the amplicons.
Restriction enzyme analysis
The restriction patterns of isolates were verified against the online database and recognized as mycobacterial species. Digestion by BstEII restriction enzyme generated three fragments of 235, 120, and 85 bp. The digestion with HaeIII enzyme also generated three fragments of 150, 130, and 70 bp. The identical restriction digestion patterns were obtained with these enzymes. Hence, PRA assay was incompetent to discriminate among MTBC. The obtained PRA patterns are shown in Figure 5.
Discussion
The Khyber Pakhtunkhwa region of Pakistan has been known for a high burden of TB and available information about identification, genotyping, and molecular basis of resistance of prevalent MTBC strains is quite limited. The current study was focused on the identification, drug resistance and genetic diversity of the existing MTBC strains found in Khyber Pakhtunkhwa, Pakistan.
Mycobacterial infections are often diagnosed by a microscopic method, which is known as smear microscopy. GeneXpert uses MTB/RIF testing to determine MTBC and rifampicin resistance in 2 h (Zeka et al., 2011). In this study, we compared the sensitivity of the fluorescence microscopy and the GeneXpert assay. Although 111 out of 794 patients tested (13.97%) were positive for MTBC by fluorescence microscopy, the positive rate was amplified to 170 (21.4%) by the GeneXpert assay. The sensitivity of the fluorescence microscopy was 74.23% while that of GeneXpert assay was 99.90%. The GeneXpert MTB/RIF test is a valuable means for managing fast diagnosis and timely treatment of TB. In a related study, conducted in Bangladesh, the MTBC-positive rate was observed to be 24.5% by fluorescence microscopy and 49.5% by GeneXpert assay (Pandey et al., 2008).
RAPD-PCR is faster and technically less demanding than most other genotyping methods. In the present study, all 52 isolates showed a high degree of polymorphisms. The level of genetic polymorphism observed was from 0 to 100%. In another similar study conducted in India, maximum discrimination was reported (Singh et al., 2006). RAPD-PCR reproducibility was found poor in a number of studies (Frothingham, 1995; Glennon and Smith, 1995). In order to avoid issues with reproducibility, the procedure was repeated three times. The high molecular diversity values resulted from RAPD PCR implied a high degree of population diversity in MTBC strains prevalent in Khyber Pakhtunkhwa, Pakistan. The existing genetic diversity among the selected population determines the variability in disease presentation, the frequency of transmission, and treatment outcomes.
In the current study, the diversity of mycobacterial clinical isolates was studied using PRA technique. The same PRA patterns observed for all MTBC clinical strains implied no usefulness of the technique. The analysis of hsp65 PRA has been reported by many other groups for identification of mycobacterial species (Häfner et al., 2004; Leão et al., 2005). A study conducted in Nigerian Institute of Medical Research (NIMR) discovered three different patterns based on the obtained number of DNA fragments and their molecular weight such as group-1 with two fragments, group–2 with one fragment and group–3 with no fragment (Nwokoye et al., 2012).
Conclusions
GeneXpert assay for identification of tuberculosis was found more sensitive than fluorescence microscopy. The studied province Khyber Pakhtunkhwa, Pakistan showed a high degree of molecular diversity based on the results from RAPD-PCR, implying a high population diversity of MTBC prevalence.
Acknowledgments
The project was supported by Higher education commission under indigenous PhD Fellowships 5000 phase II. We also acknowledge Provincial TB Reference Laboratory, Peshawar, Pakistan for providing facilities for isolation and identification of TB isolates.
Statement of conflict of interest
Authors have declared no conflict of interest.
References
Batool, B.T., Tareen, A., Ahmed, S.S., Ejaz, H., Kakar, M.A., Rehman, S. A. and Shahwani, M.N., 2017. Prevalence of zoonotic tuberculosis and brucellosis in animals of Quetta and Pishin Districts, Balochistan. Pakistan J. Zool., 49: 363-363.
Glennon, M. and Smith, T., 1995. Can random amplified polymorphic DNA analysis of the 16S-23S spacer region of Mycobacterium tuberculosis differentiate between isolates? J. clin. Microbiol., 33: 3359-3360.
Guillén-Nepita, A., Vázquez-Marrufo, G., Blanco-Guillot, F., Figueroa-Aguilar, G. and Vázquez-Garcidueñas, M., 2013. RAPD discloses high molecular diversity of Mycobacterium tuberculosis from Michoacán, Mexico. New. Microbiol., 36: 413-418.
Häfner, B., Haag, H., Geiss, H.-K. and Nolte, O., 2004. Different molecular methods for the identification of rarely isolated non-tuberculous mycobacteria and description of new hsp65 restriction fragment length polymorphism patterns. Mol. Cell. Probes., 18: 59-65. https://doi.org/10.1016/j.mcp.2003.09.003
Initiative, G.L., 2014. Mycobacteriology laboratory manual. WHO Stop TB Partnership, Geneva.
Jonsson, J., Hoffner, S., Berggren, I., Bruchfeld, J., Ghebremichael, S., Pennhag, A. and Groenheit, R.., 2014. Comparison between RFLP and MIRU-VNTR genotyping of Mycobacterium tuberculosis strains isolated in Stockholm 2009 to 2011. PLoS One, 9: e95159. https://doi.org/10.1371/journal.pone.0095159
Kim, H., Kim, S.-H., Shim, T.-S., Kim, M.-n., Bai, G.-H., Park, Y.-G., Lee, S.-H., Chae, G.-T., Cha, C.-Y. and Kook, Y.-H., 2005. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Sys. Evol. Microbiol., 55: 1649-1656. https://doi.org/10.1099/ijs.0.63553-0
Leão, S.C., Bernardelli, A., Cataldi, A., Zumarraga, M., Robledo, J., Realpe, T., Mejía, G.I., da Silva Telles, M.A., Chimara, E. and Velazco, M., 2005. Multicenter evaluation of mycobacteria identification by PCR restriction enzyme analysis in laboratories from Latin America and the Caribbean. J. Microbiol. Methods, 61: 193-199. https://doi.org/10.1016/j.mimet.2004.11.015
Marlowe, E.M., Novak-Weekley, S.M., Cumpio, J., Sharp, S.E., Momeny, M.A., Babst, A., Carlson, J.S., Kawamura, M. and Pandori, M., 2011. Evaluation of the Cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J. clin. Microbiol., 49: 1621-1623. https://doi.org/10.1128/JCM.02214-10
Nei, M. and Li, W.-H., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. natl. Acad. Sci., 76: 5269-5273. https://doi.org/10.1073/pnas.76.10.5269
Ogbaini-Emovon, E., 2009. Current trends in the laboratory diagnosis of tuberculosis. Ben. J. Postgrad. Med., 11: 79-90. https://doi.org/10.4314/bjpm.v11i1.48860
Pandey, B.D., Poudel, A., Yoda, T., Tamaru, A., Oda, N., Fukushima, Y., Lekhak, B., Risal, B., Acharya, B. and Sapkota, B., 2008. Development of an in-house loop-mediated isothermal amplification (LAMP) assay for detection of Mycobacterium tuberculosis and evaluation in sputum samples of Nepalese patients. J. med. Microbiol., 57: 439-443. https://doi.org/10.1099/jmm.0.47499-0
Siddiqi, S. and Rüsch-Gerdes, S., 2006. MGIT procedure manual. Foundation for Innovative New Diagnostics, Geneva, Switzerland, pp. 41-51.
Singh, J., Verma, R. and Chaudhuri, P., 2006. Random amplified polymorphic DNA (RAPD) analysis of Mycobacterium tuberculosis strains in India. J. Vet. Sci., 7: 181-187. https://doi.org/10.4142/jvs.2006.7.2.181
Tortoli, E., Baruzzo, S., Heijdra, Y., Klenk, H.-P., Lauria, S., Mariottini, A. and van Ingen, J., 2009. Mycobacterium insubricum sp. nov. Int. J. Sys. Evol. Microbiol., 59: 1518-1523. https://doi.org/10.1099/ijs.0.003459-0
Zeka, A.N., Tasbakan, S. and Cavusoglu, C., 2011. Evaluation of the GeneXpert MTB/RIF assay for rapid diagnosis of tuberculosis and detection of rifampin resistance in pulmonary and extrapulmonary specimens. J. clin. Microbiol., 49: 4138-4141. https://doi.org/10.1128/JCM.05434-11
To share on other social networks, click on any share button. What are these?










