Field Evaluation of Advanced Cotton Genotypes Against Insect Pest Complex in Agro-Climatic Conditions of Tandojam, Sindh
Research Article
Field Evaluation of Advanced Cotton Genotypes Against Insect Pest Complex in Agro-Climatic Conditions of Tandojam, Sindh
Muhammad Usman Asif*, Raza Muhammad, Waseem Akbar, Mubasshir Sohail and Muhammad Awais
Nuclear Institute of Agriculture, Tandojam-70060, Pakistan
Abstract | The resistance levels of eighteen cotton genotypes to sucking complex (thrips, jassids, and whiteflies) and bollworms (spotted and pink) were assessed in the experimental field of Nuclear Institute of Agriculture, Tandojam. The experiment was conducted under Randomized Complete Block Design (RCBD) with three replications. The result of overall mean revealed that NIA-M-20 had comparatively greater resistance to the attack of jassids (0.21/leaf) whitefly (0.29/leaf) and thrips (1.63/leaf). Similarly, NIA-M-16 also showed the lowest infestation of whitefly (0.29/leaf). Whereas, infestation of bollworms illustrated that NIA-M-20 was highly susceptible against spotted and pink bollworms with percent infestation of 5.52 and 2.79%, respectively. The genotype NIA-HM-27 proved to be most efficient by rendering the lowest (0.82%) infestation of spotted and pink bollworm but showed medium response and found susceptible against jassid (0.28/leaf), thrips (2.21/leaf) and whitefly (0.31/leaf). Best yield performance (2467.8 kg/ha) was of NIA-HM-6N that showed high tolerance against sucking pests as well as bollworms. Moreover, the perusal of data showed that the population trend of all insects remained below economic threshold level (ETL) during the period of study.
Received | September 14, 2018; Accepted | November 23, 2018; Published | December 12, 2018
*Correspondence | Muhammad Usman Asif, Nuclear Institute of Agriculture, Tandojam-70060, Pakistan; Email: uakhan1987@hotmail.com
Citation | Asif, M.U., Muhammad, R., Akbar, W., Sohail, M. and Awais, M. 2018. Field evaluation of advanced cotton genotypes against insect pest complex in agro-climatic conditions of Tandojam, Sindh. Journal of Innovative Sciences, 4(2): 83-89.
DOI | http://dx.doi.org/10.17582/journal.jis/2018/4.2.83.89
Keywords | Cotton genotypes, Screening, Sucking pests, Bollworms, Yield
Introduction
Cotton plays a pivotal role in agro-based economy of Pakistan and is a key source of foreign exchange earnings. It is multi-purpose crop as it provides edible oil as well as fiber. Cotton accounts for 1.9% in GDP and 8.6% of value addition in agriculture (Anonymous, 2006). In Pakistan, it is cultivated on an area of 2961 thousand hectares with 13.983 million bales and ranks 4th in overall world production after China, India and USA (GOP, 2015). The average cotton yield in Pakistan is about 752 kg/ha, which is substantially low as compared to other countries of the world (GOP, 2018). Many factors are responsible for this low productivity, but the most serious one is the intensity of insect pests attack (Arshad and Suhail, 2010).
Insect pests complex of cotton is divided into two categories; sucking pest and chewing pests or bollworms. The importat sucking insect pests are Amrasca devastans (Homopetra:Cicadellidae), Thrips tabaci (Thysanoptera:Thripidae) and Bemesia tabaci (Homoptera:Aleyrodidae). The bollworms complex includes Helicoverpa armigera (Lepidoptera:Noctuidae), Pectinophora gossypiella (Lepidoptera:Gelechiidae), Earis insulana and E. vitella (Lepidoptera:Noctuidae) (Mohyuddin et al., 1997). It is estimated that sucking and bollworms pest complex causes approximately 30-40% yield loss in Pakistan (CCRI, 2005).In order to evade these losses, farmers mostly depend on the use of pesticides (Arif et al., 2007). Major recipient of the insecticides in Pakistan is the cotton crop which receives 61.92% of the total pesticides (Khan et al., 2010).
Use of synthetic insecticides not only poses ecological contamination and health hazards but also emerging insecticidal resistance and disturbs the natural balance between the beneficial agents (pathogens, parasitoids and predators) and pests population in agro-ecosystem (Ahmad and Khan, 1991; Hamburg and Guest, 1997; Sorejani, 1998). Chandani et al. (2015) documented that environment friendly insecticides should be preferred because, conventional insecticides are mostly used to control insect pests complex but, these cause many harms i.e. resistance, resurgence, pest outbreak, damage to eco-cycle, pollution and health hazards, etc.
Host plant resistance gives insect pests control without any added cost. Moreover, this method is environment friendly and economical (Pedigo, 1989; Khan and Saxena, 1998). Insecticidal use can be minimized easily by using resistant varieties (Hua and Hua, 2000). There are several physio-morphic characters of cotton genotypes that impose various resistance/tolerance levels against the insect pest attack. In Pakistan breeders continuously made efforts to evolve insect resistant varieties by using several breeding methods, however after few years these genotypes become susceptible. According to Li et al. (2008) proper utilization of existing germplasm through hybridization and induction of new germplasm are essential to create adequate genetic variability and developing superior genotypes. Germplasm variability not only enhances the probability of developing multiple resistance against abiotic and biotic stresses but also obtain required characteristics that can be exploited in future programs of breeding (van Esbroeck and Bowman, 1998).
Keeping in view, the present study was carried out on eighteen advanced genotypes of cotton with the aim to determine their comparative attraction for sucking and chewing complexes of cotton under agro-climatic condition of Tandojam (Sindh, Pakistan) under unsprayed condition.
Materials and Methods
Total of eighteen advanced genotypes i.e. NIA-HM-2, NIA-HM-7, NIA-HM-18, NIA-HM-5, NIA-HM-20R, NIA-HM-6R, NIA-HM-30, NIA-HM-26, NIA-HM-20N, NIA-HM-27, NIA-HM-19, NIA-HM-6N, NIA-M-23, NIA-M-16, NIA-M-20, NIA-M-10, NIA-M-5 and NIA-M-9 were sown at the experimental area of Nuclear Institute of Agriculture, Tandojam on 09-05-2017. The study was conducted under RCBD with three replications. Plot size was 5m x3m with plant to plant distance 30 cm and row to row was maintained at 75 cm. All standard agronomical practices were adopted and no synthetic insecticide application was done during the whole study period.
The population of jassid, whitefly and thrips per leaf was taken early morning at fortnightly interval starting from the start of June to end of September. Three plants were randomly selected in one plot. Three leaves i.e. upper, middle and lower from each plant were observed to record the population (Ahmed et al., 2011). Bollworms infestation was recorded by observing the buds, flowers and dissecting the bolls from three plants selected randomly per plot. Percent infestation of spotted (Earis spp.) and pink (Pectinophora gossypiella) bollworms was separately calculated by recording the total and damaged number of buds, flowers and bolls from three plants per plot using the formula:
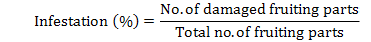
The yield from each genotype in a replicated plot was weighed and then converted in kg/ha-1. The data regarding the population of sucking insect pests, bollworms infestation and yield was analyzed by using one way ANOVA through statistix software 8.1. Means were compared through Tuckey’s HSD test. Statgraphics Centurion XVI version 16.1.11 was used for cluster analysis. Squared Euclidean distance was used for the cluster matrix. Dendrogram was developed for visualizing the results and interrelation of the cotton genotypes.
Results
The seasonal mean of jassid infestation on different cotton genotypes presented in Table 1 shows that NIA-HM-26 indicated maximum susceptibility with population of 0.54/leaf that was at par with NIA-HM-20R bearing population 0.53/leaf. The minimum population of 0.21/leaf was recorded on NIA-M-20 followed by NIA-M-23 and NIA-HM-6N with population of 0.25 and 0.26/leaf, respectively. The next best genotypes were NIA-HM-27, NIA-M-19 and NIA-HM-19 with lowest population of 0.28, 0.32 and 0.33/leaf, respectively.
The data regarding mean population of thrips on different advanced cotton genotypes is displayed in Table 2. The seasonal mean population of thrips ranged between 1.63 to 4.23/leaf among the tested genotypes. NIA-HM-20R harbored the highest (4.23/leaf) population of thrips and found as the most susceptible genotype followed by NIA-HM-6R and NIA-M-19 with population of 3.82 and 3.35/leaf, respectively. The minimum population of thrips was observed on NIA-M-20 (1.63/leaf) and found statistically at par with NIA-HM-20N bearing population 1.68/leaf. NIA-M-5, NIA-M-10 and NIA-M-23 were also found tolerant genotypes with population mean of 1.85, 207 and 2.08/leaf, respectively.
The results revealed that population of whitefly remained very low throughout the season. However, analysis of variance showed that population of whitefly varied significantly among the tested genotypes (Table 3). The highest population of 0.51/leaf was observed on NIA-HM-5 followed 0.50/leaf on NIA-HM-20R and was non-significantly different from each other. The minimum whitefly was observed on NIA-M-20 and NIA-M-16 with population mean of 0.29/leaf.
Based on the percent infestation of spotted bollworm, the results showed significant variation among different cotton genotypes (Table 4). The highest percent infestation of 7.11% was found in NIA-M-19 that
Table 1: Comparison of means of jassid population on different cotton genotypes.
|
Genotypes |
Jassid/Leaf |
Genotypes |
Jassid/Leaf |
Genotypes |
Jassid/Leaf |
|
NIA-HM-2 |
0.34 a-e |
NIA-HM-30 |
0.37 a-e |
NIA-M-23 |
0.25 de |
|
NIA-HM-7 |
0.47 a-c |
NIA-HM-26 |
0.54 a |
NIA-M-16 |
0.34 a-e |
|
NIA-HM-18 |
0.44 a-d |
NIA-HM-20N |
0.46 a-d |
NIA-M-20 |
0.21 e |
|
NIA-HM-5 |
0.42 a-e |
NIA-HM-27 |
0.28 b-e |
NIA-M-10 |
0.40 a-e |
|
NIA-HM-20R |
0.53 a |
NIA-HM-19 |
0.33 a-e |
NIA-M-5 |
0.37 a-e |
|
NIA-HM-6R |
0.48 ab |
NIA-HM-6N |
0.26 c-e |
NIA-M-19 |
0.32 a-e |
Means sharing similar letter are not significantly different at p<0.05.
Table 2: Comparison of means of thrips population on different cotton genotypes.
|
Genotypes |
Thrips/Leaf |
Genotypes |
Thrips/Leaf |
Genotypes |
Thrips/Leaf |
|
NIA-HM-2 |
3.11 a-e |
NIA-HM-30 |
2.87 b-f |
NIA-M-23 |
2.08 c-f |
|
NIA-HM-7 |
3.28 a-d |
NIA-HM-26 |
2.90 b-f |
NIA-M-16 |
2.42 c-f |
|
NIA-HM-18 |
3.29 a-d |
NIA-HM-20N |
1.68 f |
NIA-M-20 |
1.63 f |
|
NIA-HM-5 |
3.15 a-d |
NIA-HM-27 |
2.21 c-f |
NIA-M-10 |
2.07 d-f |
|
NIA-HM-20R |
4.23 a |
NIA-HM-19 |
2.40 c-f |
NIA-M-5 |
1.85 ef |
|
NIA-HM-6R |
3.82 ab |
NIA-HM-6N |
2.17 c-f |
NIA-M-19 |
3.35 a-c |
Means sharing similar letter are not significantly different at p<0.05.
Table 3: Comparison of means of whitefly population on different cotton genotypes.
|
Genotypes |
Whitefly/Leaf |
Genotypes |
Whitefly/Leaf |
Genotypes |
Whitelfy/Leaf |
|
NIA-HM-2 |
0.32 b |
NIA-HM-30 |
0.33 b |
NIA-M-23 |
0.30 b |
|
NIA-HM-7 |
0.42 ab |
NIA-HM-26 |
0.38 ab |
NIA-M-16 |
0.29 b |
|
NIA-HM-18 |
0.34 ab |
NIA-HM-20N |
0.30 b |
NIA-M-20 |
0.29 b |
|
NIA-HM-5 |
0.51 a |
NIA-HM-27 |
0.31 b |
NIA-M-10 |
0.33 b |
|
NIA-HM-20R |
0.50 a |
NIA-HM-19 |
0.44 ab |
NIA-M-5 |
0.41 ab |
|
NIA-HM-6R |
0.38 ab |
NIA-HM-6N |
0.46 ab |
NIA-M-19 |
0.42 ab |
Means sharing similar letter are not significantly different at p<0.05.
Table 4: Comparison of mean percent infestation of spotted bollworm on different cotton genotypes.
|
Genotypes |
Spotted % Infestation |
Genotypes |
Spotted % Infestation |
Genotypes |
Spotted % Infestation |
|
NIA-HM-2 |
3.59 b-d |
NIA-HM-30 |
2.11 cd |
NIA-M-23 |
4.03 a-c |
|
NIA-HM-7 |
3.51 b-d |
NIA-HM-26 |
3.82 b-d |
NIA-M-16 |
3.67 b-d |
|
NIA-HM-18 |
2.52 b-d |
NIA-HM-20N |
2.23 cd |
NIA-M-20 |
5.52 ab |
|
NIA-HM-5 |
2.37 cd |
NIA-HM-27 |
0.82 d |
NIA-M-10 |
1.72 cd |
|
NIA-HM-20R |
2.55 b-d |
NIA-HM-19 |
2.02 cd |
NIA-M-5 |
3.30 b-d |
|
NIA-HM-6R |
1.79 cd |
NIA-HM-6N |
1.03 cd |
NIA-M-19 |
7.11 a |
Means sharing similar letter are not significantly different at p<0.05.
Table 5: Comparison of mean percent infestation of pink bollworm on different cotton genotypes.
|
Genotypes |
Pink % Infestation |
Genotypes |
Pink % Infestation |
Genotypes |
Pink % Infestation |
|
NIA-HM-2 |
2.18 a-f |
NIA-HM-30 |
3.18 a-c |
NIA-M-23 |
2.53 a-e |
|
NIA-HM-7 |
2.38 a-f |
NIA-HM-26 |
3.68 a |
NIA-M-16 |
2.18 a-f |
|
NIA-HM-18 |
1.72 b-f |
NIA-HM-20N |
1.61 c-f |
NIA-M-20 |
2.79 a-d |
|
NIA-HM-5 |
2.91 a-d |
NIA-HM-27 |
0.82 f |
NIA-M-10 |
1.25 d-f |
|
NIA-HM-20R |
1.02 ef |
NIA-HM-19 |
0.85 f |
NIA-M-5 |
2.69 a-d |
|
NIA-HM-6R |
2.88 a-d |
NIA-HM-6N |
1.48 d-f |
NIA-M-19 |
3.38 ab |
Means sharing similar letter are not significantly different at p<0.05.
Table 6: Comparison of yield (kg/ha) of different cotton genotypes.
|
Genotypes |
Yield kg/ha |
Genotypes |
Yield kg/ha |
Genotypes |
Yield kg/ha |
|
NIA-HM-2 |
1741.1 a-c |
NIA-HM-30 |
1582.2 bc |
NIA-M-23 |
1282.2 c |
|
NIA-HM-7 |
1628.9 bc |
NIA-HM-26 |
1503.3 bc |
NIA-M-16 |
1216.7 c |
|
NIA-HM-18 |
1429.8 bc |
NIA-HM-20N |
1708.9 a-c |
NIA-M-20 |
1788.9 a-c |
|
NIA-HM-5 |
1643.3 bc |
NIA-HM-27 |
1723.3 a-c |
NIA-M-10 |
1693.3 a-c |
|
NIA-HM-20R |
1390.0 bc |
NIA-HM-19 |
1722.2 a-c |
NIA-M-5 |
1764.4 a-c |
|
NIA-HM-6R |
2150.0 ab |
NIA-HM-6N |
2467.8 a |
NIA-M-19 |
1525.6 bc |
Means sharing similar letter are not significantly different at p<0.05.
was statistically at par with NIA-M-20 (5.52%) and NIA-M-23 (4.03%). The most tolerant genotypes among the selected 18 cotton genotypes were NIA-HM-27 with percent infestation of 0.82% followed by NIA-HM-6N (1.03%) and NIA-M-10 (1.72%). Regarding pink bollworm infestation, NIA-HM-27 followed by NIA-HM-19, NIA-HM-20R and NIA-M-10 showed high level of tolerance with percent infestation of 0.82, 0.85, 1.02 and 1.25%, respectively. The most susceptible genotypes were NIA-HM-26, NIA-M-19, NIA-HM-30 and NIA-HM-5 having percent infestation of 3.68, 3.38, 3.18 and 2.91%, respectively, as compared with other genotypes (Table 5).
In respect of average yield per hectare, the statistically maximum yield was obtained from the genotype NIA-HM-6N i.e. 2467.8 kg /ha followed by NIA-HM-6R, NIA-M-20, NIA-M-5 with 2150, 1788.9, 1764.4 kg per hectare whereas the lowest yield was obtained in the genotype NIA-M-16 i.e. 1216.7 kg/ha followed by NIA-M-23 with 1282.2 kg/ha under similar condition (Table 6).
Cluster analysis clearly separated the genotypes into different clusters on the basis of their performance against the insect pests infestation (Fig. 1). The main cluster A that includes five genotypes viz. NIA-HM-20N, NIA-M-10, NIA-HM-27, NIA-HM-19 and NIA-HM-6N. This cluster can be divided into two sub-clusters named as AI and AII. The sub-cluster AI consisted of NIA-HM-20N and NIA-M-10 that were closely related to each other. Both of these genotypes exhibited high tolerance level against the infestation of thrips, whitefly, spotted bollworm and pink bollworm. Similarly, NIA-HM-19 and NIA-HM-6N clustered in to AII and showed least infestation of insect pests except whitefly that was found in abundance on these genotypes. However, NIA-HM-27 was found somewhat different from the other four genotypes within the same cluster. This genotype was the most tolerant with the lowest percent infestation of spotted and pink bollworms. Two genotypes, NIA-HM-2 and NIA-M-16 found closely related to each other by showing almost similar infestation of jassid and pink bollworm. While NIA-M-23, NIA-M-20, NIA-HM-18, NIA-HM-7, NIA-HM-6R, NIA-HM-30, NIA-HM-5 and NIA-HM-26 were found somewhat similar to each other in their susceptibility against insect pests and present as a separate genotype in the dendrogram. Three genotypes i.e. NIA-M-5, NIA-HM-20R and NIA-M-19 appeared to be distinctly unique and the most distant from other genotypes of cotton. NIA-M-5 was among the genotypes that showed high tolerance against the attack of thrips. While the genotype NIA-HM-20R appeared to be highly susceptible against the attack of jassid, thrips whitefly but exhibited high tolerance in case of pink bollworm attack. NIA-M-19 harbored the highest infestation of spotted bollworm and pink bollworm and regarded as the most susceptible genotype among all the tested genotypes.
Discussion
Cultivation of tolerant cotton genotypes is considered as one of the safest measures to avoid pest situation i.e., determining relative resistance in conventional genotypes, is an essential component for successful integrated pest management approach and sustainable cotton production. Eighteen advanced cotton genotypes were evaluated for their comparative resistance against insect pests complex of cotton under natural field conditions. The results revealed that tested genotypes varied significantly in their susceptibility against sucking insects of cotton. It is apparent from the results that the NIA-M-20 was the most tolerant genotype against jassid, thrips and whitefly. These findings are in conformity with Shad et al. (2001), Chu et al. (2002), Sial et al. (2003), Syed et al. (2003), Ahmad et al. (2004), Ali and Aheer (2007), Amjad et al. (2009), Khan (2011), Salman et al. (2011), Ghafoor et al. (2011), Javaid et al. (2012), Asif et al. (2017a) and Asif et al. (2017b) who also reported that different cotton genotypes varied in their relative resistance against the sucking pests. Likewise, Babar et al. (2013) conducted study on different cultivars of cotton and screened five genotypes i.e. Sitara-11M, IR5-NIBGE, Sitara-10M, Bt-121, IR4-NIBGE and Sitara-10M for resistance against jassid, thrips and whitefly. The lowest population of jassids was found on IR4-NIBGE and Sitara-11M whereas IR4-NIBGE also showed maximum tolerance against whitefly. The least susceptible variety to the infestation of thrips was Sitara-10M. Shahid et al. (2012) reported that minimum attack of thrips exhibited by FH-118, followed by GN-2085 while FH-177, FH-179 and FH-114 were most susceptible.
The present studies have indicated that attack of bollworms (spotted and pink) and yield varied substantially on the cotton genotypes. On the whole, it was observed that NIA-HM-27 exhibited least infestation of both spotted and pink bollworms. Many researchers found significant results of host plant resistance against bollworms i.e. Jin et al. (1999), Razaq et al. (2004), Nasreen et al. (2004), Pathan et al. (2007) and Lanjar et al. (2014). Solangi et al. (2016) monitored dynamics of spotted and pink bollworm on several cotton genotypes. They concluded that lowest infestation of bollworms was observed on Sattari-Bt. However, NIAB-78 also showed some tolerance against both pests.
Conclusion
The genotypes NIA-M-20 and NIA-HM-27 were found the most tolerant with respect to lowest infestation of sucking pests and bollworms, respectively. These genotypes can be included in future breeding programs for resistance enhancement and also in integrated pest management (IPM) programs for control of these pests to avoid yield losses.
Author’s Contribution
Muhammad Usman Asif planned and conducted the experiment, recorded the data, performed statistical analysis of the data and wrote the manuscript. Raza Muhammad provide technical guidance and helped in data analysis and critical review of the manuscript. Waseem Akbar contributed in execution of the experiment and helped in data collection. Mubasshir Sohail helped in data analysis and interpretation of results. Muhammad Awais helped in the manuscript writing and review.
References
Ahmad, M. and Khan, M. 1991. Insecticide resistance management strategies in cotton pests in Pakistan. Pakistan Entomologist, 13(1-2): 99-103.
Ahmad, S., Maqsood, S. Farooq, H.M.K. and Ullah, F. 2004. Resistance of cotton against Amrasca devastans (Dist.) (Jassidae: Homoptera) and relationship of the insect with leaf hair density and leaf hair length. Sarhad Journal of Agriculture, 20(2): 265-268.
Ahmad, N., Khan, M., Tofique, M. and Rauf, I. 2011. Insect pests management of Bt cotton through the manipulation of different eco-friendly techniques. The Nucleus, 48(3): 249-254.
Ali, A. and Aheer, G.M., 2007. Varietal resistance against sucking insect-pests of cotton under Bahawalpur ecological conditions. Journal of Agricultural Research, 45: 1-5.
Amjad, M., Bashir, M.H. and Afzal, M. 2009. Comparative resistance of some cotton cultivars against sucking insect pests. Pakistan Journal of Life and Social Sciences, 7(2): 144-147.
Anonymous. 2006. Federal Bureau of Statistics, Govt. of Pakistan, Ministry of Food, Agriculture & Livestock, Islamabad. pp: 12-13.
Arshad, M. and Suhail, A. 2010. Studying the sucking insect pests community in transgenic Bt cotton. International Journal of Agriculture and Biology, 12(5): 764-768.
Arif, M.J., Abbas, G. and Saeed, S., 2007. Cotton in danger, Dawn, March 24th, 2007, pp: 4. Available: URL: http://dawn.com.
Asif, M.U., Muhammad, R., Akbar, W. and Tofique, M. 2017a. Response of various cotton genotypes against sucking and bollworm complexes. Pakistan Journal of Agriculture Agricultural Engineering and Veterinary Sciences, 33(1): 37-45.
Asif, M.U., Muhammad, R., Akbar, W. and Tariq, J.A. 2017b. Performance of transgenic and non-transgenic cotton genotypes against insect pest complex. Journal of Innovative Sciences, 3(2): 45-54.
Babar, T.K., Karar, H., Hasnain, M., Shahazad, M.F., Saleem, M. and Ali, A. 2013. Performance of some transgenic cotton cultivars against insect pest complex, virus incidence and yield. Pakistan Journal of Agricultural Sciences, 50(3): 367-372.
CCRI, 2005. Annual report. Central Cotton Research Institute, Multan, Punjab, Pakistan, pp. 23.
Chandani, S. and Sathe, T. 2015. Incidence and host plants for Amrasca biguttula (ishida) from kolhapur region, India. International Journal of Development Research, 5(3): 3658-3661.
Chu, C.C., Natwick, E.T. and Henneberry, T.J. 2002. Bemisia tabaci (Homoptera: Aleyrodidae) biotype b colonization on okra-and normal-leaf upland cotton strains and cultivars. Journal of Economic Entomology, 95(4): 733-738.
GOP, 2015. Pakistan Economic Survey, 2014-15. Ministry of Finance, Government of Pakistan, 2015, pp. 23-44.
GOP, 2018. Pakistan Economic Survey, 2017-18. Ministry of Finance, Government of Pakistan, 2018, p. 15.
Ghafoor, A., Hassan, M., Alvi, Z.H. and Kausar, S. 2011. Impact of different varieties of stub cotton on population dynamics of whitefly at Faisalabad Pakistan. Pakistan Journal of Zoology, 43(1): 25-28.
Hamburg, H. and Guest, P. 1997. The impact of insecticides on beneficial arthropods in cotton agro-ecosystems in South Africa. Archives of Environmental Contamination and Toxicology, 32(1): 63-68.
Hua, M.L. and Hua, L.C. 2000. A study on the bollworm resistance of CRI- 29 and the target to control the F3 bollworms. China Cottons, 27: 20–2.
Javaid, M., Arif, M.J., Gogi, M.D., Shahid, M.R., Iqbal, M.S., Bibi, R. and Shehzad, M.A. 2012. Relative resistance in different cultivars of Pakistani cotton against cotton whitefly. Academic Journal of Entomology, 5(3): 143-146.
Jin, Z., Cao, G., Luo, S., Hong, J. and Hung, Y. 1999. Insect resistance and yield of different insect resistant hybrid cotton cultivars. Zhejiang Nongye Kexue, 3: 142-144.
Khan, Z.R. and Saxena, R.C., 1998. Host plant resistance to insects. In: Critical issues in insect pest management (eds. G.S. Dahaliwal and E.A. Heinrichs), Commonwealth Publishers, New Dehli, India, pp. 118–155.
Khan, M.J., Zia, M.S. and Qasim, M. 2010.Use of pesticides and their role in environmental pollution. World Academy of Science Engineering and Technology, 72: 122-128.
Khan, S.M., 2011. Varietal performance and chemical control used as tactics against sucking insect pests of cotton. Sarhad Journal of Agriculture, 27(2): 255-261.
Lanjar, A.G., Solangi, B.K., Khuhro S.A. and Solangi, A.W. 2014. Insect infestation on Bt and non-Bt. cotton cultivars. Food Science and Quality Management, 27: 55-62.
Li, Z., Wang, X., Zhang, Y., Zhang, G., Wu, L., Chi, J. and Ma, Z. 2008. Assessment of genetic diversity in glandless cotton germplasm resources by using agronomic traits and molecular markers. Frontiers of Agriculture in China, 2(3): 245-252.
Mohyuddin, A.I., Jilkani, G., Khan, A.G., Hamza, A., Ahmad, I. and Mahmood, Z., 1997. Integrated Pest Management of major cotton pests by Conservation, Redistribution and Augmentation of Natural Enemies. Pakistan Journal of Zoology, 29: 293-298.
Nasreen, A., Cheema, G.M., Fareed, S. and Saleem, M.A. 2004. Resistance of different cotton cultivars to chewing insect pests. Pakistan Journal of Entomology, 26(1): 81-85.
Pathan, A.K., Chohan, S., Leghari, M.A., Chandio, A.S. and Sajjad, A. 2007. Comparative resistance of different cotton genotypes against insect pest complex of cotton. Sarhad Journal of Agriculture, 23(1): 141-144.
Pedigo, L.P., 1989. Entomology and pest management. Macmillan Pub. Co., New York, pp. 413-439.
Razaq, M., Aslam, M., Shad, S.A. and Naeem, M. 2004. Evaluation of some new promising cotton strains against bollworm complex. Journal of Research (Science), 15(3): 313-318.
Salman, M., Masood, A., Arif, M., Saeed, S. and Hamed, M. 2011. The resistance levels of different cotton varieties against sucking insect pests complex in Pakistan. Pakistan Journal of Agriculture Agricultural Engineering and Veterinary Sciences, 27: 168-175.
Sial, I.A., Arif, M., Gogi, M.D. and Sial. A.A. 2003. Cataloguing of cotton genotypes for morphological traits relating to resistance against whitefly, Bemisia tabaci (Gen.). Pakistan Entomologist, 25(2): 149-153.
Shad, S.A., Waseem, A. and Rizwan, A. 2001. Relative response of different cultivars of cotton to sucking insect pests at Faisalabad. Pakistan Entomologist, 23(1/2): 79-81.
Shahid, M.R., Arif, M.J., Mahmood, A., Arshad, M., Gogi, M.D. and Elahi, F. 2012. Comparison of resistance among different cultivars of cotton against Thrips tabaci under unsprayed conditions. Pakistan Entomologist, 34 (1): 83-85.
Solangi, B., Suthar, V., Rustamani, M., Abro, N., Abbasi, A. and Abro, M. 2016. Comparative bollworm infestation on Bt and non-Bt cotton. Sindh University Research Journal (Science Series), 48(1): 23-28.
Sorejani, M., 1998. Current trend in pesticide usage in some Asian countries. Review of Applied Entomology, 77: 219–234.
Syed, T.S., Abro, G.H., Khuhro, R.D. and Dhauroo, M.H. 2003. Relative resistance of cotton varieties against sucking pests. Pakistan Journal of Biological Sciences, 6(14): 1232-1233.
Van Esbroeck, G. and Bowman, D.T. 1998. Cotton germplasm diversity and its importance to cultivar development. Journal of Cotton Science, 2: 121–129.
To share on other social networks, click on any share button. What are these?





