Enhanced Production of Streptokinase by UV- and Ethidium Bromide-Treated Streptococus equisimilis Mutant
Enhanced Production of Streptokinase by UV- and Ethidium Bromide-Treated Streptococus equisimilis Mutant
Muhammad Naeem1, Bushra Sadia1,2,*, Faisal Saeed Awan1 and Muhammad Anjum Zia3
1Centre of Agricultural Biochemistry and Biotechnology, University of Agriculture, Faisalabad 38000
2U.S.-Pakistan Center for Advanced Studies in Agriculture and Food Security, University of Agriculture, Faisalabad 38000
3Department of Biochemistry, University of Agriculture, Faisalabad 38000
ABSTRACT
Streptokinase (SK) is a thrombolytic agent having broad applications in treatment of various diseases like ischemia, Budd-Chiari syndrome, myocardial infarction, brain stroke and myocardial infarction. Strain improvement is an important approach for its hyper production. In this connection, wild bacterial strain Streptococcus equisimilis was mutagenized using ethidium bromide and UV radiation. The colonies (15 each) showing > 1% survival rate at 150 min of UV exposure and 12 % survival at 0.5 mg/ml of ethidium bromide for 120 min were selected. Casein hydrolytic assay identified EB3, EB4, EB6, EB7, EB9, EB11, EB13, UV2, UV4, UV6, UV7, UV9, UV11, UV12 and UV 15 mutants producing halo zones. Percentage blood clot lysis led to screening of mutants UV 2, UV 6, UV 7, UV 9, UV 11, UV 12, UV 15 and EB 3, EB 4, EB 11, EB 13 with hyperactivity as compared to wild bacterial strain. Out of the total 15 isolates of EtBr-treated strains, EB4 gave the maximum enzyme activity (7.3 ± 0.005 U ml-1) with a zone diameter of 6 mm. While UV6 was found to be the best UV-induced mutant with an enzyme activity of 20.333 ± 0.006A U ml-1 and 16 mm of zone diameter. The current results indicated 400 % and 146% enhanced SK activity in mutants UV6 and EB4, respectively, compared to wild type. UV6 also exhibited the highest enzyme activity (14.667± 6.5) after precipitation with ammunium sulphate, at concentrations as low as 30 μl. Optimized conditions for maximum yield of mutant SK included CSL 6%, 24 h. of fermentation, pH 7.5, 37oC, KH2PO4 0.04%, K2HPO4, NaHCO3 0.15% and 0.015% CH3COONa. The results indicated that UV is the most effective mutagen for hyper-production of SK. Mutant strain UV6 has the potential for enhanced SK production with commercial applications.
Article Information
Received 15 July 2017
Revised 06 December 2017
Accepted 13 January 2018
Available online 15 March 2018
Authors’ Contribution
BS supervised the reported research as part of PhD thesis of MN. BS, FSA and MAZ conceived the study and designed the experiments. MN performed experiments, compiled results and drafted paper. MAZ provided bacterial strains and mutation facilities. FSA supervised data analyses and edited manuscript. BS and FSA provided lab facilities.
Key words
Streptokinase, Mutagenesis, Hyper production, Precipitation, Thrombolytic agent, UV mutagenesis.
DOI: http://dx.doi.org/10.17582/journal.pjz/2018.50.2.655.661
* Corresponding author: [email protected]
0030-9923/2018/0002-0655 $ 9.00/0
Copyright 2018 Zoological Society of Pakistan
Introduction
Streptokinase (SK) is an extracellular protein secreted by various strains of streptococci. It is used clinically as an intravenous thrombolytic agent for the treatment of acute myocardial infarction (MI) by activation of plasminogen indirectly in circulation. The enzyme SK is a single chain polypeptide consisting of 414 amino acids and 47 kDa molar mass (Ganbgwar et al., 2011). SK has two domains (amino-terminal domain and carboxyl-terminal domain) which are involved in substrate recognition and activation of plasminogen (Hernandez and Marrero, 2005). Plasminogen is converted into plasmin which dissolves blood clots. Two other plasminogen activators that occur naturally in human circulatory system are the tissue type (tPA) and the urokinase type (uPA). These are used to treat blood clot within blood vessels but there is a limitation as to trigger plasminogen in absence of fibrin which results in reduction of their efficiency as drug. SK works more efficiently compared to natural plasminogen activators. It has the capacity of activation both in the absence and presence of fibrin by “catalytic switch” that contains NH2-terminal. Mutant SK lacking NH2-terminal is unable to activate plasminogen by fibrin independent mechanism (Loy et al., 2001).
The incidence of MI disease is growing at alarming rate in the world and in Pakistan (Hamid and Kalsoom, 2017). Being least expensive and more efficient, microbial sourced SK is regarded as the drug of choice for MI, compared to tPA and uPA (Ghosh et al., 2012). However, the therapeutic potential of SK is limited owing to its immunogenicity, short half-life and less quantity produced by wild bacterial strains.
The commercial applications of enzyme require its production at low cost and high reproducibility. Keeping in view the extensive applications of SK in treatment of various diseases, different strategies for enhancement in quantity and quality of SK have been investigated (Sharma et al., 2004). In this connection, strain improvement and medium optimization are two important means of hyper producing SK enzyme. Plenty of record exists on enhanced production of various industrial enzymes via strain improvement approach (Raju and Davikar, 2013). For strain improvement, mutagenesis and selection are two efficient approaches. In this regard, different physical mutagens including X-rays, Ɣ-rays, ultraviolet (UV) rays or chemical mutagens such as N-methyl-N’- nitro-N-nitrosoguanidine (MNNG), ethyl methanesulfonate (EMS) and ethidium bromide (EtBr) have been reported (Abdelghani et al., 2005; Iftikhar et al., 2010). UV and MNNG have been frequently reported as effective mutagens for strain improvement (Zia et al., 2010). UV is a preferred mutagen owing to its simplicity, effectiveness and widespread use in industry. UV light excites electrons in molecules during mutagenesis, which leads to the formation of extra bonds between adjacent pyrimidines in DNA. The UV light is the vast studied mutagenic agent in prokaryotes. Moreover, different chemical/ enzymatic and genetic modification methods have also led to the production of structurally modified SK (SK-k59E) with improved stability.
Present study describes the mutagenesis of wild S. equisimilis strain using UV and EtBr in an effort to perform strain improvement for enhanced SK production. One of the core objectives of the presented research here was to develop a baseline for hyper production of SK from bacterial strain S. equisimilis. To this end following research activities were attempted:
Materials and Methods
Mutagenesis using UV rays
Ultraviolet rays are effective mutagenic agents for strain improvement and enhanced enzyme productivity. The wild strain of S. equisimilis was subjected to mutagenesis by UV treatment (Bapiraju et al., 2004). The bacterial cell suspensions (10 ml) were transferred to sterile Petri plates and placed in UV (254 nm) chamber at a distance of 11.5 cm from UV source (Philips UVC lamp, 15W). UV lamp was warmed up to 30 min before irradiation (Irfan et al., 2011). Plates were opened during treatment to avoid shielding and covered with lids immediately after irradiation. These were kept in the black box at 37oC. The cells were subjected to UV light for different time periods ranging from 0 to 270 min at an interval of 30 min. UV irradiated cells were serially diluted in phosphate buffer and 0.1 ml of the diluted cell suspension was spread onto BHI medium and incubated at 37oC for 24 h. The number of colonies in each plate were counted after 24 h, assuming each colony originated from a single cell and colony forming units per ml were calculated. The plates were kept in a black box to avoid photo reactivation before and after irradiation. An optimum dose of 150 min was selected after preparation of the kill curve (Lin and Wang, 2001; Abdelghani et al., 2005; Devi et al., 2013).
Mutagenesis using ethidium bromide
A stock solution of ethidium bromide (0.5 mg/ ml) was prepared. One ml of the stock solution was added to 9 ml inoculum of S. equisimilis. This suspension containing ethidium bromide was incubated at 37oC. One ml of solution was drawn after a specific time interval (0, 30, 60, 90, 120, 150, 180, 210, 240 and 270 min) of incubation and was subjected to centrifugation at 10,000 rpm for 15 min, centrifugation repeated thrice for removal of the traces of mutagen (Zia et al., 2010). The kill curve established ethidium bromide dose of 120 min the best one (Devi et al., 2013). The colony forming units were calculated using following formula (Iftikhar et al., 2010):
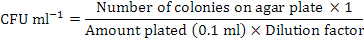
Isolation of mutants by selective marker
The mutant cells were allowed to grow in BHI supplemented with 2-deoxy-D-glucose (1 mg/ ml) at 37oC for 48 h. The mutant resistant colonies that appeared in the presence of toxic 2-deoxy-D-glucose were picked randomly and subjected to the SK identification (Iftikhar et al., 2010). Following tests were performed for the detection of hyper-producing specific mutants of S. equisimilis:
Qualitative screening of mutants
The positive strains of S. equismilis for SK were identified. The mutants producing SK were selected by caseinolytic plate method. The blood (10 ml) containing anticoagulant (EDTA) was centrifuged at 13000 rpm for 10 min. The supernatant used as plasma was mixed with nutrient agar and plated. The soft agarose overlay (10 ml) containing semi solidified agarose (0.8% agarose), 200 µl of human plasma, 10% skimmed milk, 50 mM Tris- HCl and 150 mM NaCl was poured on top of the plates. Strains were streaked and incubated at 37°C for 2-6 h. After incubation, the zones around the colonies indicated positive SK activity (Devi et al., 2013).
Selection by semi quantitative SK activity
The strains producing larger hollow zone were scratched, dissolved into phosphate buffer, filtered and subjected to enzyme activity. SK activity was determined indirectly by blood clot lysis method. The fibrinated human blood (5 ml) was taken in tubes, filtrate (100 µl) was added and incubated at 37oC for 18 h. One unit is defined as the quantity of enzyme that liquefied the blood clot completely. The strains with maximum activity were selected for quantitative screening of mutant (Devi et al., 2013).
Selection by quantitative SK activity
The strains selected by semi quantitative SK activity were subjected to enzyme production at pre-optimized conditions. Various concentrations (10, 20, 30, 40, 50, 60, 70, 80, 90 and 100 µl) of crude enzyme obtained by centrifugation at 10,000 rpm for 15 min at 0oC were used to perform enzyme assay. The strain showing maximum enzyme activity at minimum concentration was selected for further quantitative screening after partial purification by precipitation with 60 % ammonium sulfate. Finally, the strain exhibiting maximum activity after partial purification was selected for enzyme production (Srinivasan et al., 2013).
SK production
The inocula were prepared by transferring a loopful culture of S. equisimilis and its mutants in 250 ml flasks having 50 ml of sterilized inoculum medium. The corn steep liquor CSL (6%) was used as economical substrate along glucose (5%), KH2PO4 (0.04%), KH2PO4 (0.04%), CH3COONa (0.15 %) and NaHCO3 (0.15 %). The flasks were incubated for 24 h at 37oC on an orbital shaker (JEIO TEC SI-4000R, Korea) at 120 rpm (Madhuri et al., 2011; Zia et al., 2013). The conditions for fermentation were optimized using different parameters for production of SK, including pH (6, 6.5, 7, 7.5, 8, 8.5), temperature (25, 30, 35, 37, 39, 40, 45 oC), substrate concentration (CSL) (1, 2, 4, 6 and 8%), glucose (1, 3, 5, 7, 9 and 10 %), KH2PO4 (0.5, 0.2, 0.4, 0.6, 0.8, 1, 1.2 and 1.4), K2HPO4 (0.05, 0.2, 0.4, 0.6, 0.8, 1, 1.2 and 1.4), NaHCO3 (0.1, 0.15, 0.2, 0.25 and 0.45), CH3COONa (0.1, 0.15, 0.2, 0.25 and 0.45). Enzyme was harvested from liquid state fermentation by centrifugation at 10,000 rpm for 15 min at 0oC. After filtration, the residue was stored at -20oC and supernatant was assayed for enzyme activity. Biuret method was used for the estimation of protein contents of SK enzyme (Gornall et al., 1949).
Enzyme assay
The percent blood clot lysis method was used for the enzyme assay (Dubey et al., 2011).
Statistical analysis
Data obtained was analyzed by T-test, useful for assessment of means and standard error of means (Steel et al., 1997). Graphs of all recorded data were plotted on MS Excel and Statistix 8.1 software.
Results
Biochemical tests were performed for the confirmation of the bacterial strain. Clear zones were observed around the colony showing β- hemolysis in the selected strain. Long chains of Cocci with Gram positive result indicated that strains were Streptococci. The samples were inoculated on THB agar medium which is frequently used as selective medium for S. equisimilis. Catalase test was also performed; where only Staphylococcus spp. gave effervescence (positive) result. The results indicated that the observed strain was beta hemolytic; it hydrolyzed starch and exhibited negative results for VP test and catalase. The strain showed positive results for sucrose, maltose, fructose and glucose fermentation tests. These results confirmed the selected strain as S. equisimilis. After confirmation, the strain was subjected to improvement for SK production followed by screening of hyper producing strain.
Strain improvement
In the present study, SK production from S. equisimilis is improved by random mutation using UV and ethidium bromide and growing mutants under submerged fermentation conditions.
About 1000 colonies were selected for further screening. It was found that UV dose of 150 min., produced 81% killing and 19 % survival rate (2.2x103CFU ml-1) as 3 log kill of wild S. equisimilis after exposure (Fig. 1). A total of 15 colonies showing > 1% survival rate were selected from plates (150 min of UV exposure) and SK production was tested. The concentration of ethidium bromide used was 0.5 mg/ml for 120 min. which produced 88 % killing and 12 % survival rate (2.12X103CFU ml-1) as 3 log kill of wild S. euisimilis after exposure (Fig. 1).
The creation of mutant strains was followed by selection of hyper producing mutants. The selectable marker 2-deoxy-D-glucose was found effective for such selection in earlier reports (Haq et al., 2005; Zia, 2007; Adsul et al., 2007) and hence, was used in this study. Thirty resistant colonies appeared in the presence of toxic 2-deoxy-D-glucose were picked up randomly and subjected to the SK identification. The selected strains were named as EBI, EB2…….EB15 and UV1, UV2, UV3….UV15 for ethidium bromide and UV treated strain, respectively. Positive screening of SK production proceeded using casein hydrolytic assay, as earlier recommended by Devi et al. (2013) and Srinivasan et al. (2013). Strains with larger zones indicated hyper production and activity of enzyme and thus selected. Herein, mutants EB3, EB4, EB6, EB7, EB9, EB11, EB13; UV2, UV4, UV6, UV7, UV9, UV11, UV12 and UV 15 produced halo zones. Among these, mutant strains UV 2, UV 6, UV 7, UV 9, UV 11, UV 12, UV 15 and EB 3, EB 4, EB 11, EB 13 showed hyperactivity as compared to wild S. equisimilis (Table I). Out of EtBr-treated strains, maximum enzyme activity (7.3 ± 0.005 U ml-1) was observed in EB4 with a zone diameter of 6 mm, while EB9 exhibited the minimum activity (2.1333 ± 0.005 U ml-1). The mutant EB4 showed 146 % increased activity among all EB mutants (Table I). On the other hand, UV6 proved the best among all the UV mutants with an enzyme activity of 20.333 ± 0.006A U ml-1 (Table I). Overall, UV radiations proved showed a significantly better in enzyme activity as compared to EtBr (Table I). The selected hyper active mutant strains were subjected to further screening on the basis of partially purified SK enzyme activity (Table II). Screening of mutants for enzyme activity at the minimum amount of crude enzyme (30 µl) identified the strains UV6, UV9, UV11, UV12, UV15 and EB4 and EB13 with higher activity (Table III). Among all these mutants, UV6 proved the best.
Table I.- Qualitative and semi-quantitative selection of specific mutants for SK.
| Type of strain |
Zone size (mm) |
%Age clot lyses |
% Increase SK activity |
| Wild strain |
4 |
5 |
100 |
| UV6 |
16 |
20 |
400 |
| UV9 |
12 |
16 |
320 |
| UV11 |
15 |
14.8 |
296 |
| UV15 |
14 |
13 |
260 |
| UV2 |
11 |
12 |
240 |
| UV12 |
12 |
12 |
240 |
| UV7 |
8 |
7 |
200 |
| EB4 |
6 |
7.3 |
146 |
| EB13 |
6 |
7 |
140 |
| EB11 |
5 |
6 |
120 |
| EB3 |
4 |
5.1 |
102 |
Values are means of 3 Petri dishes/ mutant.
Table II.- Comparison of EtBr and UV mutants with wild bacterial strain for SK activity.
| Sr. No. |
EtBr mutagenesis |
UV mutagenesis |
||
|
Strains |
Enzyme activity Uml-1 |
Strains |
Enzyme activity Uml-1 |
|
| 1 |
EB4 |
7.3000A ± 4.5* |
UV6 |
20.333 A± 3.3** |
| 2 |
EB13 |
7.0000A± 3.6* |
UV9 |
16.000B± 6.4 |
|
3 |
EB11 |
6.0000AB± 5.2 |
UV11 |
14.833BC± 6.3* |
| 4 |
EB3 |
5.3000B± 3.2 |
UV15 |
13.067CD± 5.5* |
| 5 |
WILD |
5.0000B± 6.3 |
WILD |
5.0000EF± 4.5 |
| 6 |
EB6 |
3.1333C± 4.4 |
UV12 |
12.000D± 7.55* |
| 7 |
EB7 |
3.0000C± 5.3 |
UV2 |
12.000D± 5.7* |
| 8 |
EB9 |
2.1333C± 6.55 |
UV7 |
7.0000E± 3.5 |
| 9 |
-- |
-- |
UV4 |
3.1667F± 2.5 |
Values are means ± SD of 3 replicates/ mutant. Means followed by different letters differ significantly at P < 0.000 according to LSD all-pairwise comparison test, * represents significantly different strains by one-sided Dennett’s multiple comparisons with a control (wild).
Table III.- SK activity of S.equisimilis mutants at minimum crude SK level (30 µl).
| Sr. No. | Strains |
Enzyme activity Uml-1 |
| 1 | UV6 |
14.667A± 6.5** |
| 2 | UV11 |
9.3333 B±4.4* |
| 3 | UV9 |
8.3333 B±5.6* |
| 4 | UV15 |
8.0000 B±5.3 |
| 5 | EB13 |
8.0000 B±4.9 |
| 6 | EB4 |
7.0000 BC±6.2 |
| 7 | UV2 |
4.0000 C±6.3 |
|
8 |
UV12 |
3.6667 C±3.2 |
Values are means ± SD of 3 replicates/ strain. Means followed by the different letters differ significantly at P < 0.000 according to LSD all-pairwise comparisons test. * Represents significance according to Hsu’s multiple comparisons with the best strain.
Strains showing maximum activity at minimum amount of crude SK were selected for further screening of best mutant. Enzyme fractions harvested from UV6, UV9 and UV11 were partially purified with 60 % ammonium sulfate and SK activity of 60 % residues, supernatant and desalted samples was determined. The strain showing maximum specific activity at least concentration of SK was selected (Table IV). The strain UV6 was selected based on hyper production and activity of SK. The efficient mutant strain UV6 produced the maximum SK yield under the optimized conditions in the current research which include CSL 6%, 24 h of fermentation, pH 7.5, 37oC, KH2PO4 0.04%, K2HPO4, NaHCO3 0.15% and 0.015% CH3COONa (Fig. 2).
Table IV.- Selection of mutants based on partially purified SK.
| Strains |
SK activity (U/ml) |
Protein contents (mg/ml) |
Specific activity (U/mg) |
| UV6 |
81.222A±15.7* |
15.67A±13.8 |
10.397A±12.2 |
| UV11 |
19.778B±10.8 |
5.62B±1.8 |
6.131 B± 5.4 |
| UV9 |
14.00 B±15.1 |
4.04B±5.5 |
2.333 B± 0.6 |
| Wild |
7.88s9 B±4.2 |
3.67B±1.5 |
2.083 B± 0.45 |
Values are means ± SD of 9 replicates/ strain. Means followed by the different letters differ significantly at P < 0.000 according to LSD all-pairwise comparisons test, * is representing significance of strains by one-sided Dunnett’s multiple and Hsu’s multiple comparisons with a control (wild) and the best of SK activity.
Discussion
A range of microorganisms (Bacillus, Pseudomonas, Staphylococcus, Alteromonas, Coryne and Esherichi acoli) are known for the production of SK (Balaraman and Prabakaran, 2007). Among the Streptococci from human hosts, S. equisimilis is reported as an efficient source of SK (Babashamsi et al., 2009; Devi et al., 2013). The SK hyper production using mutagenesis was also reported earlier from different strains of S. equisimilis (Devi et al., 2013; Abdelghani et al., 2005). The present study employed bacterial strain S. equisimilis as it did not produce erythrogenic toxins. The confirmatory tests revealed that S. equisimilis was beta haemolytic, hydrolyzed starch and exhibited negative results for VP test and catalase assay, it also indicated that the strain fermented sucrose, maltose, fructose and glucose. Similar confirmatory results were observed by Madhuri et al. (2011).
Induction of random mutations under controlled conditions using chemical or physical mutagens followed by screening of improved cells is an efficient and popular method to improve microorganisms. This approach has been exploited to enhance the performance of microorganisms for monogenic and polygenic traits of commercial importance (Steensels et al., 2014). A typical mutagenesis procedure comprises of three steps: overnight growth of the bacterial strain, exposure to mutagenic agent and a recovery and screening step. In this regard, carefull selection and optimization of both the type of mutagen, the dose and exposure time should be carried out (Crook and Alper, 2012). However, the type of genetic modification brought about by any mutagen is unpredictable. Different mutagens have been exploited for strain improvement including UV, γ rays, MNNG, and ethidium bromide (Zia et al., 2013; Devi et al., 2013; Faran et al., 2015). Strain improvement in the present study involved UV and ethidium bromide (EtBr) treatments.
Our choice of UV rays as the mutagen for strain improvement is in agreement with earlier reports (Iftikhar et al., 2010; Irfan et al., 2011), where the proficiency of UV is established for enhanced production of enzymes. A low dose of the mutagen will lead to low frequency of mutants, which intern will make the screening of improved mutants difficult. Whereas, exposure of cells to higher dose of mutagen will generate large number of mutants with higher frequency of mutations, which may be beneficial or deleterious. Hence, the standardization of the optimal dose is the pre-requisite to mutagenesis approach, which typically shows a monotonic dose–response curve. The kill curve shows that the proportion of required mutants increased with increasing the mutagen dose till it reached the saturation point, proceeded by increased number of undesirable mutants. The current results indicated 400 % UV-enhanced SK activity in mutant strain UV6, compared to wild type, these results are in agreement with Devi et al. (2013), who reported increased SK production also from UV mutagenesis. Different reports exist on the effect of UV exposure time on the enhanced enzyme production from different microbes ranging from 150 min (present study) to 45 (Irfan et al., 2011) and 3 min (Adsul et al., 2007). Ethidium bromide is also reported as a potential source of chemical mutagenesis. Enhanced enzyme production has already been reported using this mutagen (Iftikhar et al., 2010; Javed et al., 2011). Present study reports 146 % increased activity (7.3 ± 0.005 U ml-1) of EtBr-induced mutant strain EB4. Similarly, Iftikhar et al. (2010) reported 124.68% increased enzyme activity using EtBr. Our results demonstrated that UV radiations showed significantly higher enzyme activity as compared to EtBr and thus were considered more proficient for mutagenesis. This finding is in line with other reports (Madhuri et al., 2011; Abdelghani et al., 2005).
Conclusion
The hunt for SK hyper producing strains is a continuous process. Present results revealed that UV and EtBR were effective mutagens for strain improvement of S. equisimilis with UV being the best mutagen producing 400 % UV-enhanced SK activity in mutant strain UV6, compared to wild type. The mutant strain has potential for commercial exploitation.
Acknowledgments
This study was conducted in the Somatic Cell Genetic Laboratory, Centre for Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture Faisalabad, Pakistan. The authors are grateful to department of Biochemistry, University of Agriculture Faisalabad, Pakistan for providing technical assistance in PhD research to MN.
Statement of conflict of interest
Authors have declared no conflict of interest.
References
Abdelghani, T.T.A., Kunamneni, A. and Ellaiah, P., 2005. Isolation and mutagenesis of streptokinase producing bacteria. Am. J. Immunol., 1: 125-129. https://doi.org/10.3844/ajisp.2005.125.129
Adsul, M.G., Bastawde, K.B., Varma, A.J. and Gokhale, D.V., 2007. Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresour. Technol., 98: 1467-1473. https://doi.org/10.1016/j.biortech.2006.02.036
Babashamsi, B.M., Razavian, M.H. and Nejadmoghaddam, M.R., 2009. Production and purification of streptokinase by protected affinity chromatography. Avicenna J. med. Biotechnol., 1: 47-51.
Balaraman, K. and Prabakaran, G., 2017. Production and purification of a fibrinolytic enzyme (thrombinase) from Bacillus sphaericus. Indian J. med. Res. 126:459–64.
Bapiraju, K.V.V.S.N., Sujatha, P., Ellaiah, P. and Ramana, T., 2004. Mutation induced enhanced biosynthesis of lipase. Afri. J. Biotechnol., 3: 618-621.
Crook, N. and Alper, H.S., 2012. Classical strain improvement. Engineering complex phenotypes in industrial strains (ed. R. Patnaik). John Wiley and Sons, Inc., Hoboken, NJ, pp. 1-33. https://doi.org/10.1002/9781118433034.ch1
Devi, C.S., Mohanasrinivasan, V., Vaishnavi, B., Selvarajan, E. and Naine, S.J., 2013. Optimization studies for enhanced production of streptokinase by S. equisimilis UVM6. J. Pure appl. Microbiol., 7: 2337-2341.
Dubey, R., Kumar, J., Agrawala, D., Char, T. and Pusp, P., 2011. Isolation, production, purification, assay and characterization of fibrinolytic enzymes (nattokinase, streptokinase and urokinase) from bacterial sources. Afri. J. Biotechnol., 10: 1408-1420.
Essam, K., 2012. Fibrinolytic bacterial enzymes with thrombolytic activity. Springer Briefs in microbiology. Springer-Verlag, Berlin, Heidelberg. eBook ISBN: 978-3-642-24980-8. Available at: http://www.springer.com/gp/book/9783642249792 (Accessed on 14 January, 2018).
Faran, G., Zia, M.A., Shahid, M. and Sajid, A., 2015. Improved streptokinase production; UV irradiation of Streptococcus equisimilis. Profess. med. J., 22: 656-663.
Ganbgwar, D.S., Lakhera, P.C. and Rai, A., 2011. Restriction enzyme analysis of recombinant plasmids containing streptokinase gene. Biotechnol. Int., 4: 50-55.
Ghosh, M., Pulicherla, K.K., Rekha, V.P.B., Rao, G.V. and Rao, K.R.S.S., 2012. A review on successive generations of streptokinase based thrombolytic agents. Int. J. Pharm. Pharmacuet. Sci., 3: 975-1491.
Gornall, A.G., Bardwill, C.J. and David, M.M., 1949. Determination of serum proteins by means of biuret reagent. J. biol. Chem., 177: 751-766.
Hamid, A. and Kalsoom, S., 2017. Comparative analysis of nutritional composition and effect of dietary fiber extracts of chickpea and Bengal gram on blood glucose and cholesterol levels of male induced diabetic and hypercholesterolemic rats. Pakistan J. Zool., 49: 487-492.
Haq, I., Ali, S., Qadeer, M.A. and Iqbal, J., 2005. Optimization of nitrogen for citric acid productivity by a 2-deoxy D-glucose resistant culture of Aspergillus niger NGd-280. Bioresour. Technol., 96: 645-648. https://doi.org/10.1016/j.biortech.2004.06.010
Hernandez, L. and Marrero, M.A., 2005. Streptokinase about of a thrombolytic patented in Cuba. Biotecnol. Appl., 22: 191-198.
Iftikhar, T., Niaz, M., Abbas, S.Q., Zia, M.A., Ashraf, I., Lee, V. and Haq, I., 2010. Mutation induced enhanced biosynthesis of lipases by Rhizopus oligosporus var. Microsporus. Pak. J. Bot., 42: 1235-1249.
Irfan, M., Javed, J. and Syed, Q., 2011. UV mutagenesis of Aspergillus niger for enzyme production in submerged fermentation. Pak. J. Biochem. mol. Biol., 44: 137-140.
Javed, M.A., Haq, I.U. and Mariyam, 2011. Multistep mutagenesis for the over-expression of cellulase in humicola insolens. Pakistan J. Bot. 43:669-677.
Lin, K. and Wang, A., 2001. UV mutagenesis in Escherichia coli K-12 Cell survival and mutation frequency of the chromosomal genes lacZ, rpoB, ompF, and ampA. J. exp. Microbiol. Immunol., 1:32-46.
Loy, J.A., Lin, X., Schenone, M., Castellino, F.J., Zhang, X.C. and Tang, J., 2001. Domain interactions between streptokinase and human plasminogen. Biochemistry, 40: 14686-14695. https://doi.org/10.1021/bi011309d
Madhuri, D.H, Manohar, M., Singh, N.A., Mohanasrinivasan, V. and Devi, C.S., 2011. Studies on isolation, screening and strain improvement of streptokinase producing β-hemolytic Streptoococci. World J. Sci. Technol., 1: 7-11.
Raju, E.V.N. and Divakar, G., 2013. Production of pectinase by using Bacillus circulans isolated from dump yards of vegetable wastes. Int. J. Pharmaceut. Sci. Res., 4: 2615-2622.
Sharma, S., Texeira, A., Texeira, P., Elias, E., Wilde, J. and Olliff, S.P., 2004. Pharmacological thrombolysis in Budd Chiari syndrome: A single centre experience and review of the literature. J. Hepatol., 40: 172-180. https://doi.org/10.1016/j.jhep.2003.09.028
Srinivasan, M.V., Devi, C.S., Saha, A., Mondal, M., Dhar, S., Suganthi, V. and Selvasrajan, E., 2013. Screening for staphylokinase producing Staphylococcus spp. from bovine milk sample. Int. J. Pharm. Pharm. Sci., 5: 601-604.
Steel, R.G.D., Torrie, J.H., Dickey, D.A., 1997. Principle and procedure of statistics. A biochemetrical approach, 3rd ed. McGraw Hill Book Co. Inc., New York.
Steensels, J., Snoek, T., Meersman, E., Nicolino, M.P., Voordeckers, K. and Verstrepen, K.J., 2014. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev., 38: 947-995. https://doi.org/10.1111/1574-6976.12073
Zia, M.A., 2007. Mutagenesis of A. niger for hyper production of glucose oxidase to prepare glucose diagnostic kit. Dissertation, University of Agriculture, Faisalabad, Pakistan.
Zia, M.A, Rahman, K., Sheikh, M.A. and Khan, I.A., 2010. Chemically treated strain improvement of Aspergillus niger for enhanced production of glucose oxidase. Int. J. Agric. Biol., 12: 153-154.
Zia, M.A., Faisal, R., Abbas, R.Z., Faran, G.E., Saleemi, M.K. and Khan, J.A., 2013. Comparison of streptokinase activity from Streptococcus mutants using different substrates. Pak. Vet. J., 33: 77-79.
To share on other social networks, click on any share button. What are these?










