Comparative Genetic Analysis for Yield and Quality Traits in Flue Cured Tobacco
Comparative Genetic Analysis for Yield and Quality Traits in Flue Cured Tobacco
Qaizar Ahmed1, Fida Mohammad2, Sheraz Ahmed2*, Sultan Akbar Jadoon2, Imtiaz Ali2 and Ajmalud Din2
1Pakistan Tobacco Board, Peshawar, Khyber Pakhtunkhwa, Pakistan; 2Department of Plant Breeding and Genetics, The University of Agriculture, Peshawar, 25130, Khyber Pakhtunkhwa, Pakistan.
Abstract | Precise knowledge regarding genetic architecture of production traits is important to devise effective breeding and selection strategies. This investigation was carried out to elucidate relationship between Hayman and Griffing analyses in flue cured Virginia (FCV) tobacco. Plant material comprised of seven tobacco cultivars which were hybridized in full diallel mating system at the Tobacco Research Sub-station, Mansehra, Pakistan during 2007. Thereon, seven parents and their forty-two F1 hybrids were sown during 2008 and 2009 at the Tobacco Research Station, Mardan (plain) and the Tobacco Research Sub-station, Mansehra (hilly). Locations in each year were considered as independent environments: Mardan 2008 as environment-1; Mansehra 2008 as environment-2; Mardan 2009 as environment-3 and Mansehra 2009 as environment-4. Field trials were conducted using randomized complete block design in four replicates. Data for all traits were subjected to Griffing and Hayman analyses of variance to get critical comparison of the results obtained through both models. Griffing analysis revealed additive type of gene action in the expression of all traits across all environments, except nicotine content. Contrarily, Hayman analysis identified both (additive and dominance) type of gene actions for yield and other traits. Similarly, Hayman analysis discriminated gene actions for various traits more specifically across environments. Based on this study, Griffing analysis was revealed to be more accommodative, whereas Hayman analysis was more stringent in specifying gene actions for various traits.
Received | September 19, 2018; Accepted | February 28, 2019; Published | April 26, 2019
*Correspondence | Sheraz Ahmed, Department of Plant Breeding and Genetics, The University of Agriculture, Peshawar, 25130, Khyber Pakhtunkhwa, Pakistan; Email: drsheraz@aup.edu.pk
Citation | Ahmed, Q., F. Mohammad, S. Ahmed, S.A. Jadoon, I. Ali and A. Din. 2019. Comparative genetic analysis for yield and quality traits in flue cured tobacco. Sarhad Journal of Agriculture, 35(2): 500-512.
DOI | http://dx.doi.org/10.17582/journal.sja/2019/35.2.500.512
Keywords | Flue cured Virginia, Tobacco, Genetic analysis, Hayman, Griffing, Additive gene action, Nicotiana tabacum
Introduction
Flue Cured Virginia tobacco is commonly known as cigarette tobacco. It is cultivated in very localized areas because of its unique growth and development requirements. In Pakistan, annual area allocation for tobacco is about 35000 ha which produces about 80 million kg. Tobacco breeders have been striving for improving its yield and quality related traits to make it more valuable for tobacco industries. Knowledge of gene action is a pre-requisite for improving any trait (Dhanda et al., 2002). Diallel analysis could be a very useful and effective technique when identifying gene actions (Gardner and Eberhart, 1966). Various procedures have been proposed to deduce information from diallel mating. Mostly used procedures are (Hayman, 1954a; 1954b), Gardner and Eberhart (1966) and (Griffing 1956a; 1956b), in which the effects and the square sum of effects for general and specific combining ability are estimated. Griffing’s analysis extracts information regarding gene action by manipulating effects due to general combining ability (GCA) and specific combining ability (SCA), whereas Hayman’s analysis is based on estimation of components of variance. However, the most frequently used methods are Griffing, Gardner and Eberhart (Gardner and Eberhart, 1966; Griffings, 1956a). Because their analyses are relatively simple, easy and very accommodative in terms of plant material. Unlike Hayman analysis, Griffings method is equally feasible in inbred lines, clones, self-pollinated and cross-pollinated species. In contrast, Hayman’s method also includes graphical array of variances and covariances along with statistical analysis, hence is able to estimate various genetic attributes (Schuelter et al., 2010). The genetic interpretation of parameters and relationship between Griffing and Hayman’s analyses have been discussed earlier by Farshadfar et al. (2012) in wheat. The purpose of this study was to evaluate and compare two diallel mating models, viz., Griffing’s and Hayman’s analyses, with regard to their combinatory effects for seven FCV tobacco genotypes.
Materials and Methods
Forty-two F1 hybrids were developed from seven tobacco varieties of distinct characters (Table 1) at the Tobacco Research Sub-station, Mansehra (runs under Pakistan Tobacco Board) during 2007-08. Seven parents and their F1 progenies were planted during 2008-09 at respective research stations of Mardan (plain) and Mansehra (hilly) following randomized complete block design in four replications. Experiments conducted at two locations during two years (2 × 2) were referred to as four distinct environments i.e environment-1 (Mardan during 2008), environment-2 (Mansehra during 2008), environment-3 (Mardan during 2009) and environment-4 (Mansehra during 2009). Description of each experimental site is presented in Table 2. Each entry of experimental material was planted in a 1-row plot of 6 m length. Plant to plant distance was 60 cm, while row to row spaces were maintained at 90 cm. Standard cultural practices were followed as recommended for tobacco crop.
Data collection
At maturity, data were detailed for plant height by measuring from the ground level to the tip after topping; for leaf area as proposed by Suggs et al. (1960); for cured leaves weight by weighing cured leaves in a plot, for yield by converting cured leaves weight into kg ha-1, for grade index by visual assessment of cured leaves, for nicotine and reducing sugar contents following Cundiff and Markunas (1964). Preceding to diallel analysis, pooled analyses of variance were computed using SAS software (SAS, 2009) to check the impact of interaction due to genotype by environment (GEI) on various traits (Table 3). In case of significant GEI, analysis of variance over each environment were performed, using appropriate model for RCB design following Steel and Torrie (1980).
Griffing analysis of variance
Significant data obtained after ANOVA were subjected to diallel analysis according to Griffing’s (1956a) Method-I based on Eisenhart’s Model-II as described by Singh and Chaudhary (1985). Diallel models were analysed using DIALLEL computer software (Burow and Coors, 1994).
Hayman analysis of variance
Genetic components of variations were estimated following Hayman’s approach of diallel analysis as explained by Singh and Chaudhry (1985).
Testing the validity of hypothesis for Hayman’s analysis
To assess validity of data for additive dominance model, three scaling tests (regression analysis, arrays analysis of variance ‘Wr + Vr’ and ‘Wr – Vr’ and the t2 test) were employed. Data of a trait will be valid for genetic interpretation if the regression coefficient is significantly different from zero but not from unity (Mather and Jinks, 1982). Failure of this test concludes the presence of epistasis and the data are deemed to be unfit for genetic interpretation. The significance of Wr + Vr array analysis shows the presence of non-allelic interactions and if there exists epistasis, Wr – Vr will vary between arrays. Similarly, if the t2 test is non-significant, this will confirm the absence of non-allelic interaction hence, genes will be considered independent in their action for random association. The model is declared fully adequate when all the tests assumptions are supported, the model is declared partially adequate if at least one test fulfils the assumptions. The model is totally unfit if none of the tests fulfils the assumptions. The t2 test validated the model for all traits except nicotine content in environment-1, environment-2 and environment-4 and leaf area and nicotine content in environment-3, where the value of t2 test was found
Table 1: Seven tobacco varieties and their characteristics.
| S. No. | Variety/line | Pedigree | Main features |
| 1 | NC 606 | NC729/NC 82 |
Good cured-leaf quality Tall plants with about 30 leaves and longer internodal length. |
| 2 | K 399 | (Coker139x Coker319) and NC 95 | Dwarf plants with about 26 leaves. Somewhat late flowering. Broader leaves. |
| 3 |
Spt. G 126
|
K 326 x Speight G-96 |
Produces average yields of less than average quality Produces nearly 20 to 25 leaves on a stalk of average height Flowers later than most varieties and good holding ability. |
| 4 | Spt. G 28 | (Coker 139 x Oxford 1-181) and NC 95 |
The recommended variety for last 30 years in Pakistan. Moderate yield and medium quality tobacco. Short plants having more than 25 leaves. Flowers medium to late. |
| 5 | KHG 21 | Locally selected accession | Based on long-term data it has tall plants, more than 30 leaves per plant and intermediate internodal length. It has more leaf area than KHG 22and KHG 24. |
| 6 | KHG 22 | Locally selected accession | Based on long-term data, it possesses an average of 24 leaves per plant with intermediate leaf area. It has more internodal length than KHG21 and KHG 24. |
| 7 | KHG 24 | Locally selected accession | Based on long-term data, it has an average of 25 leaves per plant with intermediate internodal length. It is dwarf, having smaller leaf area than KHG21 and KHG 22. |
Table 2: Description of the experimental environments.
| E-1 | E-2 | E-3 | E-4 | ||
| Altitude (meters) | 283 | 975.36 | 283 | 975.36 | |
| Annual Rainfall (mm) | 480.98 | 926.00 | 275 | 605.50 | |
| Rainfall during March to August (mm) | 339.68 | 524.50 | 114 | 388.00 | |
| Mean annual temperature (Min and Max oC) | 13.4-30.1 | 13.3-27.5 | 15.5-32.6 | 13.0-27.8 | |
| Mean temperature, March to August (Min and Max oC) | 18.1-35.1 | 17.8-32.5 | 18.9-36.2 | 16.9-32.4 | |
| Soil texture | Silt loam | Silt loam | Silt loam | Silt loam | |
| Soil chemical nature | pH | 7.70-7.83 | 7.2 | 7.78 | 7.2 |
| Chlorides % | 0.001 | 0.006 | 0.002 | 0.006 | |
| Soil nutrients | N % | 0.04-0.08 | 0.185 | 0.048 | 0.179 |
| P % | 0.0008 | 0.00098 | 0.001 | 0.00096 | |
| K % | 0.0115 | 0.018 | 0.0115 | 0.0174 | |
| Organic matter % | 0.6 | 2.1 | 0.6 | 2.0 | |
significant inferring presence of epistasis. According to the parameters employed on data for adequacy of Additive-Dominance model, all the traits were found partially adequate in all 4 environments except cured leaf weight plot-1 and yield in environment-1 and grade index in environment-4 which were found fully adequate. However, it was found not adequate for nicotine content in all four environments.
Hayman’s graphical analysis
In Hayman’s analysis, gene action is concluded by plotting the co-variances (Wr) of each array against its variance (Vr). The slope and position of the regression line fitting to the array points within the limiting parabola indicates the degree of dominance and the presence/absence of gene interaction. The points of limiting parabola (Wri) were obtained using the formula:
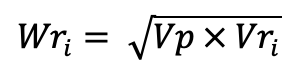
Where;
Vp = paternal variance and Vri = array variance.
The distance between the origin and the point where the regression line intersects the Wr-axis provides a measure of average degree of dominance;
Partial dominance: (D > H1); Complete dominance: (D = H1); Over dominance: (D < H1) and the regression line touches parabola limit is an indication of no dominance.
Table 3: Mean squares for plant height, leaf area, cured leaves weight, yield, grade index, nicotine and reducing sugars in 7×7 diallel of FCV tobacco, during 2008 and 2009.
| Source | df | PH | LA | CW | YLD | GI | NC | RS |
| Combined ANOVA | ||||||||
| Environment | 3 | 16664.4** | 17446049.21** | 1887112.4** | 188711246.1** | 2080.7** | 30.27** | 27.6** |
| Rep (Env.) | 12 | 65.5 | 73869.60 | 9851.7 | 985170.4 | 31.3 | 0.32 | 5.8 |
| Genotype | 48 | 54.2** | 35269.68** | 2319.2** | 231918.3** | 193.6** | 0.76** | 69..3** |
| G x E | 144 | 46.2** | 20280.99** | 1875.3** | 187534.1** | 44.5** | 0.01** | 2.7** |
| Error | 576 | 27.6 | 10079.94 | 656.1 | 65608.0 | 14.9 | 0.007 | 0.151 |
| CV % | 5.02 | 10.08 | 8.49 | 8.49 | 5.38 | 3.88 | 2.04 | |
| E-1 | ||||||||
| Block | 3 | 158.49** | 231405.37** | 277.89 | 27788.29 | 73.23** | 0.008 | 0.34 |
| Genotype | 48 | 30.88 | 46547.29** | 1497.21** | 149720.86** | 75.03** | 0.192** | 18.11** |
| Error | 144 | 22.72 | 13781.32 | 157.44 | 15743.95 | 13.34 | 0.006 | 0.18 |
| CV % | 4.35 | 8.53 | 3.06 | 3.06 | 5.33 | 4.37 | 2.16 | |
| E-2 | ||||||||
| Block | 3 | 31.47 | 60263.79 | 30668.33** | 3066832.48** | 36.80 | 1.161** | 18.59** |
| Genotype | 48 | 80.28* | 42313.96** | 2935.84** | 293584.52** | 79.09** | 0.239** | 20.65** |
| Error | 144 | 51.54 | 23548.28 | 1398.75 | 139874.94 | 17.84 | 0.008 | 0.19 |
| CV % | 7.59 | 14.11 | 10.54 | 10.54 | 6.06 | 3.58 | 2.27 | |
| E-3 | ||||||||
| Block | 3 | 50.98 | 1299.57 | 500.67 | 50066.82 | 3.23 | 0.019* | 0.67** |
| Genotype | 48 | 65.83** | 4031.52** | 1279.52** | 127951.84** | 77.57** | 0.187** | 19.88** |
| Error | 144 | 26.09 | 1657.47 | 232.42 | 23242.27 | 13.50 | 0.006 | 0.14 |
| CV % | 4.45 | 5.62 | 7.73 | 7.73 | 4.86 | 4.35 | 1.99 | |
| E-4 | ||||||||
| Block | 3 | 21.16 | 2659.01 | 7959.94** | 795994.18** | 11.50 | 0.110** | 3.50** |
| Genotype | 48 | 15.95* | 3186.07** | 2232.64** | 223263.48** | 95.31** | 0.171** | 18.79** |
| Error | 144 | 10.04 | 1331.96 | 835.71 | 83570.70 | 15.00 | 0.007 | 0.10 |
| CV % | 3.19 | 4.59 | 11.82 | 11.82 | 5.29 | 3.48 | 1.66 |
* and ** ; P ≤ 0.05 and P≤ 0.01, respectively; E-1: Mardan 2008; E-2: Mansehra 2008; E-3: Mardan 2009; E-4: Mansehra 2009; PH: plant height; LA: leaf area; CW: cured leaves weight; YLD: yield; GI: grade index, NC: nicotine and RS: reducing sugars.
Results and Discussion
Analysis of variance (ANOVA)
Significant differences were detected among seven FCV tobacco varieties along with their 42 F1 hybrids for various attributes. Interaction due to genotype by environment was also significant (P<0.01) for all the studied traits suggesting genetic sensitivity of genotypes to varying environments. Hence, it demanded the consideration of analysis in each environment. Significant differences were noticed among genotypes for all studied attributes in four environments (Table 3) which justified genetic analyses for further evaluation.
Griffing analysis of variance
Identification of superior parental lines with desirable traits is an important prerequisite for hybridization program to generate vigorous genotypes. Similarly, information regarding additive and non-additive gene action is of utmost importance for a plant breeder to efficiently determine the progress of cultivar development program (Farshadfar et al., 2012). Such information could help in screening of parental combinations for identifying populations with potential segregants.
Combining ability is actually the capability of a genotype to inherit superior performance to its progenies. The efficiency of an inbred line is determined by the its ability to yield superior hybrids when involved in a series of crosses. For suitable parental selection,
Table 4: Mean performance of parents for plant height, leaf area, cured leaves weight, yield, grade index, nicotine and reducing sugars in 7×7 diallel of FCV tobacco, during 2008 and 2009.
| PH | LA | CW | YLD | GI | NC | RS | |
| (cm) |
(cm2) |
(g) |
(kg ha-1) |
(%) | (%) | (%) | |
| E-1 | |||||||
| KHG22 | 111.0 | 1407 | 422 | 4218 | 64.4 | 1.42 | 17.0 |
| Spt G 126 | 108.8 | 1314 | 450 | 4500 | 78.4 | 1.62 | 18.5 |
| K399 | 106.5 | 1359 | 420 | 4199 | 73.3 | 1.44 | 22.2 |
| NC 606 | 105.5 | 1324 | 373 | 3731 | 73.1 | 1.34 | 17.9 |
| KHG21 | 106.0 | 1194 | 404 | 4037 | 64.1 | 1.49 | 20.3 |
| KHG24 | 110.5 | 1274 | 391 | 3912 | 67.4 | 1.79 | 17.3 |
| Spt G 28 | 107.8 | 1271 | 397 | 3972 | 78.1 | 1.66 | 20.4 |
| Grand mean | 108.0 | 1306 | 408 | 4081 | 71.3 | 1.54 | 19.1 |
|
Lsd (P<0.05) |
6.7 | 164 | 18 | 175 | 5.1 | 0.11 | 0.6 |
| E-2 | |||||||
| KHG22 | 95.9 | 1266 | 391 | 3915 | 65.1 | 2.11 | 15.7 |
| Spt G 126 | 87.5 | 938 | 329 | 3286 | 80.0 | 2.02 | 17.4 |
| K399 | 93.9 | 1051 | 341 | 3414 | 74.6 | 1.68 | 22.4 |
| NC 606 | 91.2 | 999 | 364 | 3636 | 74.7 | 2.38 | 18.3 |
| KHG21 | 89.9 | 917 | 354 | 3539 | 67.8 | 2.13 | 19.8 |
| KHG24 | 92.0 | 978 | 325 | 3250 | 69.8 | 2.49 | 19.5 |
| Spt G 28 | 90.1 | 971 | 415 | 4150 | 79.6 | 2.34 | 20.1 |
| Grand mean | 91.5 | 1017 | 360 | 3598 | 73.1 | 2.16 | 19.0 |
|
Lsd (P<0.05) |
10.0 | 215 | 52 | 523 | 5.9 | 0.12 | 0.6 |
| E-3 | |||||||
| KHG22 | 121.5 | 716 | 169 | 1694 | 63.8 | 1.41 | 15.5 |
| Spt G 126 | 110.3 | 737 | 209 | 2093 | 85.8 | 1.61 | 18.1 |
| K399 | 118.0 | 734 | 225 | 2252 | 74.8 | 1.44 | 21.5 |
| NC 606 | 114.6 | 676 | 195 | 1949 | 81.0 | 1.35 | 17.1 |
| KHG21 | 116.4 | 773 | 240 | 2404 | 71.8 | 1.48 | 19.3 |
| KHG24 | 119.4 | 720 | 191 | 1911 | 76.8 | 1.75 | 16.8 |
| Spt G 28 | 112.5 | 730 | 197 | 1967 | 80.3 | 1.78 | 20.1 |
| Grand mean | 116.1 | 727 | 204 | 2039 | 76.3 | 1.54 | 18.3 |
|
Lsd (P<0.05) |
7.1 | 57 | 21 | 213 | 5.1 | 0.11 | 0.5 |
| E-4 | |||||||
| KHG22 | 100.7 | 769 | 229 | 2289 | 64.8 | 2.12 | 15.4 |
| Spt G 126 | 102.0 | 820 | 249 | 2492 | 79.9 | 2.32 | 17.6 |
| K399 | 97.9 | 819 | 220 | 2203 | 71.8 | 2.13 | 19.6 |
| NC 606 | 100.9 | 862 | 272 | 2722 | 73.1 | 2.05 | 19.8 |
| KHG21 | 103.8 | 791 | 265 | 2646 | 66.5 | 2.19 | 19.4 |
| KHG24 | 100.8 | 799 | 202 | 2019 | 65.7 | 2.45 | 16.4 |
| Spt G 28 | 100.8 | 748 | 299 | 2994 | 81.3 | 2.48 | 22.0 |
| Grand mean | 100.9 | 801 | 248 | 2481 | 71.9 | 2.25 | 18.6 |
|
Lsd (P<0.05) |
4.4 | 51 | 40 | 404 | 5.4 | 0.12 | 0.4 |
PH: plant height; LA: leaf area; CW: cured leaves weight; YLD: yield; GI: grade index, NC: nicotine and RS: reducing sugars; E-1: Mardan 2008; E-2: Mansehra 2008; E-3: Mardan 2009; E-4: Mansehra 2009.
combining ability analysis is one the most effective methods in determining the genetic value of inbred lines (Singh and Narayanan, 1993; Singh and Chaudhary, 1999; Chapi et al., 2008; Mohammadi et al., 2010). In the current study, the existed genetic variability for each trait was apportioned into combining ability and reciprocal effects following Sprague and Tatum (1942); Griffing (1956), respectively. Many researchers opined that GCA effects involve additive type of gene activity, whereas non-additive type of gene activities is governed by SCA effects. However, the concept that GCA is a sign of additive gene action seems to be disputed among researchers (Matzinger, 1963). Jugenheimer (1976) believed that the present information regarding the aforementioned concept was insufficient and urged the need of more experiments to prove its validity. Mean squares for GCA were significantly different for all traits in all four environments (Table 5). For SCA, ANOVA revealed significant variation for plant height, leaf area, cured leaves weight, yield, grade index, nicotine and reducing sugars studied at environment-1 and for cured leaf weight, yield, grade index, nicotine and reducing sugars at environment-2. Similarly, SCA effects were significant for grade index, nicotine and reducing sugars at environment-3 and for grade index, nicotine and reducing sugars at environment-4. According to Cruz et al. (2004), the presence of significant variance amongst GCA effects (gi) is deemed to be responsible for additive gene effects, whereas SCA effects (Sij) are associated with non-additive gene effects. It is also apparent from Table 5 that GCA values are greater in magnitude than those for SCA indicating the predominance of additive gene effects in controlling the heredity of all the traits studied. The improvement through selection in such case would therefore, be simple and speedy. Baker (1978) and Cisar et al. (1982) proposed that the ratio of combining ability variance components [(2σ2GCA)/ (2σ2GCA + σ2SCA)] was helpful in predicting the performances of progeny. High ratio (close to unity) offers greater opportunity of realistic estimation based on GCA alone. Some researchers have a view that GCA effects are steady and last longer in self-crossing populations. On the contrary, SCA is generation dependant which fluctuates as the generation advances and is predominantly driven by the type of gene action concerned that governs a character (Masood and Kronstad, 2000). This could be explained by the fact that dominant and epistatic gene actions decrease with the advancement of generations, due to decreasing trend in the frequency of heterozygous individuals in the population.
Table 5: Mean squares for combining ability for traits measured from 7-line FCV tobacco diallel cross in 4 environments, during 2008 and 09.
| Df | PH | LA | CW | YLD | GI | NC | RS | ||
| E-1 | GCA | 6 | 14.55* | 27442.30** | 1409.8** | 140982.7** | 43.36** | 0.026** | 5.48** |
| SCA | 21 | 10.16* | 13282.08** | 145.3** | 14527.6** | 13.79** | 0.058** | 3.35** | |
| Reciprocal | 21 |
3.32 ns |
5475.72 ns |
307.6** | 30764.2** | 16.70** | 0.044** | 5.43** | |
|
Error' |
144 | 5.68 | 3445.33 | 39.4 | 3935.3 | 3.33 | 0.001 | 0.04 | |
| E-2 | GCA | 6 | 31.75* | 41154.35* | 1877.4** | 187735.3** | 32.49** | 0.060** | 6.88** |
| SCA | 21 |
17.84 ns |
6962.26 ns |
706.7* | 70665.4* | 18.17** | 0.073** | 3.43** | |
| Reciprocal | 21 |
18.96 ns |
5458.76 ns |
434.6 ns |
43461.4 ns |
17.74** | 0.046** | 6.41** | |
|
Error' |
144 | 12.88 | 5887.07 | 349.7 | 34965.8 | 4.45 | 0.002 | 0.05 | |
| E-3 | GCA | 6 | 43.25** | 4238.37** | 1336.8** | 133680.4** | 118.66** | 0.025** | 6.51** |
| SCA | 21 |
7.63 ns |
486.84 ns |
82.8 ns |
8281.7 ns |
9.68** | 0.055** | 3.53** | |
| Reciprocal | 21 | 17.63** |
605.92 ns |
266.3** | 26629.7** |
0.75 ns |
0.044** | 5.97** | |
|
Error' |
144 | 6.52 | 414.37 | 58.1 | 5809.4 | 3.37 | 0.001 | 0.04 | |
| E-4 | GCA | 6 | 8.29** | 3135.04** | 1553.9** | 155392.0** | 123.98** | 0.022** | 5.85** |
| SCA | 21 |
3.26 ns |
415.56 ns |
318.3 ns |
31828.6 ns |
13.06** | 0.049** | 2.85** | |
| Reciprocal | 21 |
3.49 ns |
509.32 ns |
513.7** | 51370.1** | 5.98ns | 0.042** | 6.22** | |
|
Error' |
144 | 2.51 | 332.99 | 208.9 | 20890.0 | 3.75 | 0.002 | 0.02 |
* : P < 0.05 and ** : P < 0.01; PH: plant height; LA: leaf area; CW: cured leaves weight; YLD: yield; GI: grade index, NC: nicotine and RS: reducing sugars; E-1: Mardan 2008; E-2: Mansehra 2008; E-3: Mardan 2009; E-4: Mansehra 2009.
Table 6: Estimation of components of genotypic variance for traits measured at Environment-1 (Mardan 2008).
| Components | PH | LA | CW | YLD | GI | NC | RS |
| D | -1.54 | Not adequate | 581.1 | 58109.1 | 32.78 | Not adequate | 3.59 |
|
+ 4.11 |
+ 43.2 |
+ 4320.2 |
+ 2.36 |
+ 1.04 |
|||
|
H1 |
9.26 | 229.3 | 22926.1 | 26.58 | 9.19 | ||
|
+ 9.88 |
+ 104.0 |
+ 10400.7 |
+ 5.69 |
+ 2.51 |
|||
|
H2 |
9.39 | 210.6 | 21061.2 | 20.30 | 6.62 | ||
|
+ 8.71 |
+ 91.6 |
+ 9164.5 |
+ 5.01 |
+ 2.21 |
|||
| F | -3.70 | 208.4 | 20835.2 | 27.71 | 4.61 | ||
|
+ 9.85 |
+ 103.6 |
+ 10364.0 |
+ 5.67 |
+ 2.50 |
|||
|
h2 |
7.05 | 6.1 | 607.0 | 28.47 | 0.80 | ||
|
+ 5.85 |
+ 61.6 |
+ 6155.3 |
+ 3.37 |
+ 1.49 |
|||
| E | 6.27 | 40.0 | 3997.0 | 3.64 | 0.05 | ||
|
+ 1.45 |
+ 15.3 |
+ 1527.4 |
+ 0.84 |
+ 0.37 |
|||
|
(H1/D)1/2 |
2.45 | 0.63 | 0.63 | 0.90 | 1.60 | ||
|
H2/4H1 |
0.25 | 0.23 | 0.23 | 0.19 | 0.18 | ||
|
(4DH1)1/2 + F / (4DH1)1/2 – F |
0.34 | 1.80 | 1.80 | 2.77 | 2.34 | ||
| Heritability (ns) | 10.54 | 67.87 | 67.87 | 39.44 | 31.36 | ||
| Heritability (bs) | 34.90 | 86.14 | 86.14 | 74.71 | 98.17 |
Parameter value is significant when it exceeds 1.96 after dividing it with its standard error; PH: plant height; LA: leaf area; CW: cured leaves weight; YLD: yield; GI: grade index, NC: nicotine and RS: reducing sugars.
According to Topal et al. (2004), in contrast to dominant gene effects, high frequency of additive gene action for a particular attribute can boost the chances of success through selection in that trait.
Hayman analysis
Environment-1 (Mardan 2008): Magnitude of dominance components H1 and H2 were greater than that of additive component D for reducing sugar and
plant height which indicated an important role of dominance variance (Table 6). However, negative value of D revealed the absence of additive gene effects in plant height. Significant variance due to additive D and dominance H components for cured leaves weight, yield and grade index indicated the involvement of both additive and dominant gene effects in the heredity of these traits. The values of H1 and H2 were nearly equal for plant height and cured leaves weight indicating presence of positive and negative genes in equal frequencies. This phenomenon was also confirmed by the ratio of H2/4H1 having value nearly equal to 0.25 for these traits. The value of E was significant for plant height, cured leaves weight, yield and grade index which indicated influence of environment on these traits. Component h2 was significant (Table 6) for grade index showing importance of dominant effects due to overall heterozygous loci. Mean degree of dominance for yield was 0.63 (smaller than 1), which signalled towards incomplete dominance. The regression line (Figure 1A) also uncovered the same phenomenon as it was having positive intercept over origin. Position of array points implicated the presence of dominant genes in parental cultivars K399 and KHG21, whereas recessive genes were the driving force in genetic makeup of NC606. Generally, heritability estimates were high in broad sense (86%) as well as in narrow sense (68%).
Environment-2 (Mansehra 2008): The additive component D was positive and significant (Table 7), whereas the dominance components H were non-significant for leaf area, cured leaves weight and yield implying the importance of additive type of gene action over dominance in these traits (Table 7). On the contrary, both additive D and dominant H components were found positive and significant for grade index and reducing sugars (Table 7) which confirmed the existence of both additive and dominant gene effects in the heredity of these traits. However, additive (D) and dominance components (H) were non-significant for plant height showing the absence of both types of gene action (Table 7) which could be the result of environmental effect. The values of H1 and H2 were close for plant height indicating the equal proportion of negative and positive alleles, while unequal values of H1 and H2 for the rest of the traits indicated asymmetrical occurrence of negative and positive alleles in the parents which was confirmed by the deviation of ratio H2/4H1 from its expected value of 0.25. Great proportion of recessive genes for leaf area was evident by negative value of F and smaller ratio of dominant and recessive genes (under unity). Partial dominance was suggested for leaf area, cure weight and yield on the basis of average degree of dominance over loci (H1/D)½. Mean degree of dominance was more than 1 for plant height, grade index and reducing sugar showing presence of over dominant genes in the breeding material for these traits. The ratio of dominant to recessive genes for cured weight, yield, grade index and reducing sugar was greater than one indicating the important role of dominant genes in the inheritance of these traits. For yield, smaller value of average degree of dominance (<1) signalled towards partial dominance. Similarly, placement of array points (Figure 1B) inferred the presence of dominant genes in parental cultivars KHG24 and KHG22, whereas recessive genes were the predominant force in the genetic makeup of Spt G 126. Estimates of narrow sense heritability were generally low for the studied traits due to smaller number of genes which acted additively.
Environment-3 (Mardan 2009): The breeding material under study exhibited additive gene effects in the inheritance of all studied traits as the D component was positive and significant but as H1 and H2 parameters were also significant for grade index and reducing sugar; therefore, both additive and dominant gene actions were involved for these two traits (Table 8). The frequency of dominant and recessive alleles in the parents suggested smaller proportion of dominant genes for plant height, leaf area and reducing sugars which was confirmed by the ratio of dominant to receive genes in parent cultivars which was not equal to 0.25 however, grade index and yield showed significant F value which pointed towards greater frequency of dominant genes in parents for these traits which was supported by higher ratio of dominant to recessive genes. Yield and grade index were environment driven traits as suggested by the
Table 7: Estimation of components of genotypic variance for traits measured at Environment-2 (Mansehra 2008).
| Components | PH | LA | CW | YLD | GI | NC | RS |
| D | -5.16 | 7791.40 | 604.8 | 60479.9 | 28.59 |
Not Adequate |
4.52 |
|
+ 3.01 |
+ 999 |
+ 230.8 |
+ 23075.5 |
+ 3.12 |
+ 0.80 |
||
|
H1 |
10.81 | 1181.25 | 456.1 | 45609.7 | 36.92 | 9.28 | |
|
+ 7.26 |
+ 2406 |
+ 555.5 |
+ 55553.5 |
+ 7.50 |
+ 1.93 |
||
|
H2 |
10.12 | 1775.73 | 415.3 | 41530.2 | 27.24 | 6.57 | |
|
+ 6.39 |
+ 2120 |
+ 489.5 |
+ 48950.4 |
+ 6.61 |
+ 1.70 |
||
| F | -9.89 | -2825.92 | 251.8 | 25177.9 | 30.29 | 5.31 | |
|
+ 7.23 |
+ 2397 |
+ 553.6 |
+ 55357.5 |
+ 7.47 |
+ 1.92 |
||
|
h2 |
33.07 | 16823.52 | -146.0 | -14601.8 | 43.93 | -0.05 | |
|
+ 4.29 |
+ 1424 |
+ 328.8 |
+ 32877.4 |
+ 4.44 |
+ 1.14 |
||
| E | 12.78 | 6074.39 | 499.0 | 49900.3 | 4.55 | 0.14 | |
|
+ 1.07 |
+ 353 |
+ 81.6 |
+ 8158.4 |
+ 1.10 |
+ 0.28 |
||
|
(H1/D)1/2 |
1.45 | 0.39 | 0.87 | 0.87 | 1.14 | 1.43 | |
|
H2/4H1 |
0.23 | 0.38 | 0.23 | 0.23 | 0.18 | 0.18 | |
|
(4DH1)1/2 + F / (4DH1)1/2 – F |
0.20 | 0.36 | 1.63 | 1.63 | 2.75 | 2.39 | |
| Heritability (ns) | 15.04 | 43.47 | 24.62 | 24.62 | 26.00 | 35.07 | |
| Heritability (bs) | 29.08 | 47.32 | 37.60 | 37.60 | 70.35 | 94.86 |
Parameter value is significant when it exceeds 1.96 after dividing it with its standard error; PH: plant height; LA: leaf area; CW: cured leaves weight; YLD: yield; GI: grade index, NC: nicotine and RS: reducing sugars.
Table 8: Estimation of components of genotypic variance for traits measured at Environment-3 (Mardan 2009).
| Components | PH | LA | CW | YLD | GI | NC | RS |
| D | 8.72 | 430.38 | 489.78 | 48978.5 | 47.98 |
Not adequate |
4.35 |
|
+ 2.59 |
+ 204 |
+ 22.18 |
+ 2218.3 |
+ 1.66 |
+ 1.13 |
||
|
H1 |
-1.83 | -26.60 | 26.94 | 2694.2 | 14.67 | 9.48 | |
|
+ 6.23 |
+ 492 |
+ 53.40 |
+ 5340.5 |
+ 4.00 |
+ 2.73 |
||
|
H2 |
1.96 | 148.60 | 46.72 | 4672.0 | 12.71 | 6.98 | |
|
+ 5.49 |
+ 433 |
+ 47.06 |
+ 4705.7 |
+ 3.52 |
+ 2.40 |
||
| F | -5.52 | -837.92 | 105.05 | 10505.1 | 16.98 | 5.01 | |
|
+ 6.20 |
+ 490 |
+ 53.22 |
+ 5321.7 |
+ 3.98 |
+ 2.72 |
||
|
h2 |
3.74 | -179.06 | 148.78 | 14877.7 | 0.21 | 1.84 | |
|
+ 3.68 |
+ 291 |
+ 31.61 |
+ 3160.6 |
+ 2.37 |
+ 1.61 |
||
| E | 6.65 | 412.54 | 59.46 | 5945.7 | 3.32 | 0.04 | |
|
+ 0.91 |
+ 72 |
+ 7.84 |
+ 784.3 |
+ 0.59 |
+ 0.40 |
||
|
(H1/D)1/2 |
0.46 | 0.25 | 0.23 | 0.23 | 0.55 | 1.48 | |
|
H2/4H1 |
-0.27 | -1.40 | 0.43 | 0.43 | 0.22 | 0.18 | |
|
(4DH1)1/2 + F / (4DH1)1/2 – F |
0.18 | -0.59 | 2.68 | 2.68 | 1.94 | 2.28 | |
| Heritability (ns) | 42.28 | 54.86 | 71.95 | 71.95 | 71.71 | 34.13 | |
| Heritability (bs) | 46.23 | 58.59 | 76.56 | 76.56 | 85.54 | 98.58 |
Parameter value is significant when it exceeds 1.96 after dividing it with its standard error; PH: plant height; LA: leaf area; CW: cured leaves weight; YLD: yield; GI: grade index, NC: nicotine and RS: reducing sugars.
Table 9: Estimation of components of genotypic variance for traits measured at Environment-4 (Mansehra 2009).
| Components | PH | LA | CW | YLD | GI | NC | RS |
| D | 0.46 | 1031.71 | 882.3 | 88229.1 | 41.49 |
Not adequate
|
5.07 |
|
+ 0.68 |
+ 95 |
+ 61.7 |
+ 6165.1 |
+ 2.09 |
+ 0.79 |
||
|
H1 |
0.52 | 195.47 | 337.8 | 33781.6 | 21.61 | 7.16 | |
|
+ 1.65 |
+ 229 |
+ 148.4 |
+ 14842.2 |
+ 5.04 |
+ 1.91 |
||
|
H2 |
1.38 | 151.61 | 146.0 | 14601.9 | 18.64 | 5.62 | |
|
+ 0.95 |
+ 202 |
+ 130.8 |
+ 13078.1 |
+ 4.44 |
+ 1.68 |
||
| F | -2.04 | 276.92 | 700.2 | 70018.9 | 10.10 | 4.95 | |
|
+ 1.64 |
+ 228 |
+ 147.9 |
+ 14789.8 |
+ 5.02 |
+ 1.91 |
||
|
h2 |
7.97 | -41.79 | -72.1 | -7214.7 | 5.58 | -0.02 | |
|
+ 0.98 |
+ 135 |
+ 87.8 |
+ 8783.8 |
+ 2.98 |
+ 1.13 |
||
| E | 2.57 | 339.76 | 245.3 | 24527.6 | 3.74 | 0.04 | |
|
+ 0.24 |
+ 34 |
+ 21.8 |
+ 2179.7 |
+ 0.74 |
+ 0.28 |
||
|
(H1/D)1/2 |
1.06 | 0.44 | 0.62 | 0.62 | 0.72 | 1.19 | |
|
H2/4H1 |
0.67 | 0.19 | 0.11 | 0.11 | 0.22 | 0.20 | |
|
(4DH1)1/2 + F / (4DH1)1/2 – F |
-0.35 | 1.89 | 4.58 | 4.58 | 1.41 | 2.40 | |
| Heritability (ns) | 21.94 | 51.39 | 39.88 | 39.88 | 67.17 | 36.48 | |
| Heritability (bs) | 31.19 | 56.27 | 47.67 | 47.67 | 85.39 | 98.18 |
Parameter value is significant when it exceeds 1.96 after dividing it with its standard error; PH: plant height; LA: leaf area; CW: cured leaves weight; YLD: yield; GI: grade index, NC: nicotine and RS: reducing sugars.
significance of their environmental components. Although, average degrees of dominance for reducing sugars pointed towards over-dominance but the positive intercept of regression line (Fig not shown) over-ruled this phenomenon. Average degree of dominance for yield was smaller than unity suggested incomplete dominance also backed by regression line of WrVr graph (Figure 1C). From the placement of array points for yield, it was implied that plenty of dominant genes were involved in NC606 and Spt
Table 10: Comparison of Griffing and Hayman’s approach in inferring gene action for different traits in FCV tobacco across four environments.
| Trait | Griffing's approach | Hayman's approach | ||||||
| E-1 | E-2 | E-3 | E-4 | E-1 | E-2 | E-3 | E-4 | |
| Plant height | Additive | Additive | Additive | Additive | Non-additive | Both | Additive | Not clear |
| Leaf area | Additive | Additive | Additive | Additive | x | Additive | Additive | Additive |
| Cured weight plot-1 | Additive | Additive | Additive | Additive | Both | Additive | Additive | Both |
| Yield | Additive | Additive | Additive | Additive | Both | Additive | Additive | Both |
| Grade index | Additive | Additive | Additive | Additive | Both | Both | Both | Both |
| Nicotine contents | Non-additive | Non-additive | Non-additive | Non-additive | x | x | x | x |
| Reducing sugars | Additive | Additive | Additive | Additive | Non-additive | Both | Both | Both |
x: Here model was not adequate for analysis of gene action; E-1: Mardan 2008; E-2: Mansehra 2008; E-3: Mardan 2009; E-4: Mansehra 2009.
G126, whereas majority of genes in KHG21 were recessive. Magnitude of narrow sense heritability was moderate for all traits suggested insufficiency of additive genes in the inheritance of the studied traits.
Environment-4 (Mansehra 2009): The significant values of additive component in leaf area, cured leaves weight, yield, grade index and reducing sugars suggested additive type of inheritance in the expression of these traits, but as H components were also significant for cured leaves weight, yield, reducing sugars and grade index, hence concurrent effect of H and D gene action is involved in the parents for these traits (Table 9). The pattern of gene action was unknown in plant height due to non-significant values of D and H components. Influence of environment for plant height, leaf area, yield and grade index were evident due to significance of environmental variance.
Plant height and flag leaf area revealed negative and non-significant frequency of dominant and recessive alleles in the parents (Table 9). The ratio of dominant to recessive genes in the parents was lesser than 1 for these traits, while the rest of traits possess significant F value directing towards significant part of dominant genes in the appearance of these character. Partial dominance was observed in controlling the yield character as mean value of dominance (0.62) was less than 1. In Wr, Vr graph the allocation of array point (Figure 1D) depicts the cultivars, Spt G 126 and KHG21 as holder of maximum dominant genes for yield, whereas Spt G 28 had maximum number of recessive genes.
Additive gene action was predominant in the expression of yield in tobacco in environment-2 and environment-3, while both additive and dominant gene effects were present in environment-1 and environment-4 (Table 10). Therefore, early generation selection will be fruitful in environment-2 and environment-3. However, selection efficiency is also related to the magnitude of heritability (Manal, 2009). According to Legg and Collins (1975); Mehta et al. (1985) and Sadeghi et al. (2012), tobacco yield was dominated by additive gene effects. However, Legg and Collins (1970) and Butorac et al. (1999) reported dominant gene effects in controlling tobacco yield. Partial dominance was revealed by average degree of dominance and graphic representation of regression line in environments (Figure 1) but contrary to findings in this study, Butorac et al. (2004) reported over-dominance type of gene action in the expression of yield.
Griffing versus Hayman approach
Hayman model is based on estimation of variance components, whereas Griffing model relies on the significance of combining ability effects. Hayman model extracts information through analysing six components, i.e., D, H1, H2, E, F, and h2, while Griffing model obtains information regarding D and H components through GCA and SCA variances. The Hayman analysis could work out genetic ratios, while it was not possible with the Griffing approach. Griffing analysis was based on the data of progeny irrespective of data for parental cultivars, while Hayman analysis could not be carried out without taking into account the data for parental cultivars. Griffing analysis identified superior cross combinations, while the Hayman analysis indicated gene actions but did not identify specific cross combinations with desirable performance. The Hayman analysis was able to extracted a lot more genetic information than Griffing analysis from the same set of data (Table 10). The contrasting results of the two studied methods for identifying gene action could be attributed to multiple factors such as the statistical procedure of estimating various parameters and the proportion of assumptions satisfied in each model. Theoretically, the experimental material should constitute the entire population about which valid inferences have to be made but practically this is impossible and reliance on sampling has to be made. In sampling assumptions, it is necessary to determine whether the experimental material is truly representing the whole population or not (Griffing, 1956a). While determining degree of dominance, Gardner (1963) indicated that in early generations the values tended to be overestimated because of upward bias due to repulsion phase of linkage. In later generations, the linkage was broken due to recombination and a low degree of dominance was obtained. In the present study, early generation (F1) was used to compute gene actions for various traits. Perusal of Table 3 indicates the difference in the amount of rainfall received across various environments. In environment-2, the higher performance could be assigned mainly to high rainfall in that environment (Table 4). Plant samples used for taking data notes was a fraction of the entire population and might not have been a true representative of the environment. Some of these factors and use of different methods of analysis tended to create differences in gene actions. For example, additive in one method of analysis and non-additive in other method analysis within in the same environment for a particular trait could happen. Hence, the estimates of heritability mostly vary across locations and years even in the same populations.
Conclusions and Recommendations
In the current study, Griffing analysis revealed additive gene actions in the inheritance of all traits across all environments, except nicotine content which was predicted to be under non-additive genes control across all four environments. However, the Hayman analysis recognized varying gene actions with more specificity and partitioning various traits across environments. Therefore, the Griffing analysis appeared less discriminative compared to Hayman analysis for identifying gene actions. Based on this study, it can be concluded that the Hayman’s analysis appeared very stringent and was best in indicating the gene actions with more specificity, whereas Griffing analysis was accommodative in predicting gene expressions.
Acknowledgements
The authors would please to acknowledge Pakistan Tobacco Board staff and workers for endowment of their assets and services during this research. The outcome of this research paper would have not been realized without the assistance of Pakistan Tobacco Board.
Author’s Contribution
Qaizar Ahmed (QA) and Fida Mohammad conceived the research. QA conducted the research. Sheraz Ahmed helped in data collection and wrote the first draft with Sultan Akbar Jadoon. Imtiaz Ali and Ajmalud Din made corrections and formatted the first draft.
References
Baker, R.J. 1978. Issues in diallel analysis. Crop Sci. 18: 533–536. https://doi.org/10.2135/cropsci1978.0011183X001800040001x
Burow, M.D. and J.G. Coors. 1994. Diallel: A microcomputer program for the simulation and analysis of diallel crosses. Agron. J. 86: 154-158. https://doi.org/10.2134/agronj1994.00021962008600010028x
Butorac, J. 1999. Inheritance of some leaf parameters in burley tobacco. Agric. Conspectus Sci. 64(2): 87-96.
Butorac, J., J. Beljo and J. Gunjaca. 2004. Study of inheritance of some agronomic and morphological traits in burley tobacco by graphic analysis of diallel cross. Plant Soil Environ. 50(4): 162–167. https://doi.org/10.17221/4078-PSE
Chapi, O.G., A.S. Hashemi, E. Yasari and G.A. Nematzadeh. 2008. Diallel analysis of seedling traits in canola. Int. J. Plant Breed. Genet. 2(1): 28-34. https://doi.org/10.3923/ijpbg.2008.28.34
Cisar, C., C.M. Brown and H. Jedlinski. 1982. Diallel analysis for tolerance in winter wheat to the barley yellow dwarf virus. Crop Sci. 22: 328–333. https://doi.org/10.2135/cropsci1982.0011183X002200030009x
Cruz, C.D., A.J. Regazzi and P.C.S. Carneiro. 2004. Modelos biométricos aplicados ao melhoramento genético. Univ. Federal de Viçosa, Viçosa. p. 480.
Cundif, R.H. and P.C. Markunas. 1964. Determination of alkaloids. Tob. Sci. 8: 136.
Dhanda, S.S., G.S. Sethi and K.K. Behl. 2002. Inheritance of seedling traits under drought stress conditions in bread wheat. Cereal Res. Commun. 30(3-4): 293-300.
Farshadfar, E., F. Rafiee and A. Yghotipoor. 2012. Comparison of the efficiency among half diallel methods in the genetic analysis of bread wheat (Triticum aestivum L.) under drought stress condition. Ann. Biol. Res. 3 (3):1607-1622.
Gardner, C.O. 1963. Estimates of genetic parameters in cross-fertilizing plants and their implications in plant breeding. p. 225-252. In: Statistical genetics and plant breeding. W.D. Hanson and H.F. Robinson (eds.), NAS-NRC Publ. 982.
Gardner, C.O. and S.A. Eberhart. 1966. Analysis and interpretation of the variety cross diallel and related populations. Biometrics. 22: 439- 452. https://doi.org/10.2307/2528181
Griffing, B. 1956a. Concept of general and specific combining ability in relation to diallel crossing systems. Aust. J. Biol. Sci. 9: 463-493. https://doi.org/10.1071/BI9560463
Griffing, B. 1956b. A generalized treatment of the use of diallel crosses in quantitative inheritance. Heredity. 10: 31-50. https://doi.org/10.1038/hdy.1956.2
Hayman, B.I. 1954(a). The analysis of variance of diallel crosses. Biometrics. 10: 235-244. https://doi.org/10.2307/3001877
Hayman, B.I. 1954(b). The theory and analysis of diallel crosses. Genetics. 39: 789-809.
Jugenheimer, R.W. 1976. Corn: Improvement, seed production, and uses. Wiley interscience, New York. 156.
Legg, P.D. and G.B. Collins. 1970. Genetic parameters in burley populations of tobacco (Nicotiana tabacum L.) I. ‘Ky 10’ x ‘Burley 21’1. Crop Sci. 11(3): 365-367. https://doi.org/10.2135/cropsci1971.0011183X001100030016x
Legg, P.D. and G.B. Collins. 1975. Genetic parameters in aky 14 xky ex 42 burley population of Nicotiana tabacum L. Theo. App. Gen. 45(6): 264-267. https://doi.org/10.1007/BF00831899
Masood, M.S. and W.E. Kronstad. 2000. Combining ability analysis over various generations in a diallel cross of bread wheat. J. Agric. Res. 16: 1-4.
Mather, K. and J.L. Jinks. 1982. Biometrical genetics. 3rd ed. Chapman and hall London, UK. https://doi.org/10.1007/978-1-4899-3406-2
Mehta, A., G.J. Patel and B.G. Jaisani. 1985. Genetic analysis of some agro-morphological traits of Nicotiana tabacum L. Tob. Res. 11(2): 148.
Mohammadi, A.A., G. Saeidi and A. Arzani. Genetic analysis of some agronomic traits in flax (Linum usitatissimum L.). Aust. J. Crop Sci. 4(5): 343-352.
Sadeghi, S.M., E. Amin and M. Ashouri. 2012. An investigation of gene action on different traits of tobacco under irrigated and drought stress environment. Afr. J. Biotech. 11(21): 4740-4751. https://doi.org/10.5897/AJB11.1525
SAS Institute Inc. 2009. SAS/STAT ® 9.2 User’s Guide, Second Edition. Copyright © 2009, SAS Inst. Inc., Cary, NC, USA. (https://support.sas.com/documentation/cdl/en/statug/63033/HTML/default/vie wer.htm)
Schuelter, A.R., G.M. Pereira, A.T. Amaral Júnior, V.W.D. Casali, C.A. Scapim, W.S. Barros and F.L. Finger. 2010. Genetic control of agronomically important traits of pepper fruits analyzed by Hayman’s partial diallel cross scheme. Genet Mol. Res. 9 (1): 113-127. https://doi.org/10.4238/vol9-1gmr694
Singh, P. and S.S Narayanan. 1993. Biometrical techniques in plant breeding. 1st Ed. Kalayani publishers, New Delhi.
Singh, R.K. and B.D. Choudhry. 1999. Biometrical methods in quantitative genetic analysis. Kalyani Publ. Ludhiana, New Delhi, revised Ed. p. 123.
Singh, R.K. and B.D. Chaudhary. 1985. Biometrical methods in quantitative genetic analysis. Kalyani Publ. Ludhiana New Delhi. pp. 102-164.
Sprague, G.F. and L.A. Tatum. 1942. General and specific combining ability in single crosses of corn. J. Am. Soc. Agron. 34: 923-932. https://doi.org/10.2134/agronj1942.00021962003400100008x
Suggs, P.W., J.E. Beeman and W.E. Splinter. 1960. Physical properties of green Virginia types tobacco leaves in relation to length and breadth of leaf area. Tob. Sci. 4: 194-197.
Topal, A., C. Aydin, N. Akgün and M. Babaoglu. 2004. Diallel cross analysis in durum wheat (Triticum durum Desf.): Identification of best parents for some kernel physical fitures. Field Crop. Res. 87(1): 1-12. https://doi.org/10.1016/j.fcr.2003.08.015
Valério, I.P., F.I.F. de Carvalho, A.C. de Oliveira, V.Q. de Souza, G. Benin, D.A.M. Schmidt, G. Ribeiro, R. Nornberg and H. Luch. 2009. Combining ability of wheat genotypes in two models of diallel analysis. Crop Breed. Appl. Biotech. 9: 100-107. https://doi.org/10.12702/1984-7033.v09n02a01
To share on other social networks, click on any share button. What are these?








