Research Journal for Veterinary Practitioners
Research Article
Prevalence and Molecular Detection of Intimin (eaeA) Virulence Gene in E. coli O157:H7 in Calves
Afaf Abdulrahman Yousif*, Mohammed Ali Hussein
Department of Internal and Preventive veterinary Medicine, College of Veterinary Medicine, University of Baghdad, Iraq.
Abstract | This study was carried out to investigate the prevalence of Escherichia coli O157:H7 serotype from diarrheic and non-diarrheic calves. The study was out in Baghdad, a province in Iraq. A total of 350 faecal samples from 35 diarrheic calves and 315 non- diarrheic calves with different ages (up to 1 year) and from both sexes. After initially enrichment, samples were streaked on sorbitol MacConkey agar plus cifixime potassium tellurite (SMA-CT) and Chrom agar™ E .coli O157:H7. Non-sorbitol fermenting (NSF) E. coli isolates were conducted to serotyping using commercial Latex agglutination test for detection of O157 and H7 antigen. E. coli Isolates were additionally tested for virulence factor eae by PCR techniques. Four isolates (11.42%) belonged to E. coli O157:H7 in 35 diarrheic calves and 28 (8.88%) in non- diarrheic calves. All four isolates from diarrheic calves were found positive for intimin (eaeA) gene (100%) and only 13 from 28 isolates (46.42%) were possessing (eaeA) gene in none diarrheic calves. In conclusion, this study revealed the importance of calves to act as a reservoir for E. coli O157:H7. Also, the eaeA genes in a high percentage in most calves suggest that they may be virulent for humans.
Keywords | E. coli O157:H7, Intimin (eaeA) gene, PCR, E. coli O157:H7 in calves
Editor | Muhammad Abubakar, National Veterinary Laboratories, Islamabad, Pakistan.
Received | May 20, 2015; Revised | June 04, 2015; Accepted | June 05, 2015; Published | June 12, 2015
*Correspondence | Afaf Abdulrahman Yousif, University of Baghdad, Iraq; Email: [email protected]
Citation | Yousif AA, Hussein MA (2015). Prevalence and molecular detection of intimin (eaeA) virulence gene in E. coli O157:H7 in calves. Res. J. Vet. Pract. 3(3): 47-52.
DOI | http://dx.doi.org/10.14737/journal.rjvp/2015/3.3.47.52
ISSN | 2308-2798
Copyright © 2015 Yousif and Hussein. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Enterohemorrhagic Escherichia coli (EHEC) strains, of which E. coli O157:H7 is the best-studied serotype, Shiga toxin-producing Escherichia coli O157:H7 causes foodborne infections, and cattle are the primary reservoir which harbour the bacteria in their intestinal tracts without showing clinical symptoms (Kieckens et al., 2015; Katani et al., 2015).
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 responsible for frequent haemorrhagic colitis and haemolytic uremic syndrome in humans. In 1982 E. coli O157:H7 was first recognized as a human pathogen (Riley et al., 1982). As it was associated with consumption of undercooked ‘hamburgers’. As it has been found that healthy cattle can harbour the bacterium, ruminants are now regarded as its main reservoir, though STEC O157:H7 has been isolated from other animal species such as pigs, sheep, geese, gulls, geese and pet animals (Gyles, 2007).
The ability of the organism to survive in feed, water, soil and manure has important implications for its persistence in cattle herds and contamination of water supplies and crops. Effective measures to reduce or eliminate E. coli O157:H7 in cattle will reduce not only food borne illness but also the risk of transmission of the organism into the environment (Bach et al., 2002).
A wide range of prevalence estimates ranging from 0.1% to 62% of E. coli O157 in cattle was reported worldwide (Lin et al., 2001; Reinstein et al., 2009; Pennington, 2010). The inconsistent prevalence estimates of E. coli O157 reported in cattle in various geographical locations might be, to some extent, due to variable methodological modus operandi to identify the organism, such as sampling strategy, type of samples, enrichment procedures, immunomagnetic separation and cultural media of choice. Therefore, the factors that contribute to the variability in the detection of the organism and thus in the prevalence estimate need to be identified by analysing the available published reports (Islam et al., 2014).
The “top five” EHEC serotypes are defined as E. coli strains harbouring Shiga toxin (stx) and intimin (eae) genes and belonging to one of the following serotypes:O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 (ANSES, 2010). Intimin (eaeA gene) and Tir (tir gene) are key colonization factors, which paly significant roles in E. coli O157:H7attachment to host epithelium (McNeilly et al., 2010; Zhang et al., 2014).
Blanco et al. (2004) recorded that Intimin is required for intimate bacterial adhesion to epithelial cells inducing a characteristic histopathological lesion defined as “attaching and effacing” (A/E). This lesion is governed by a large pathogenicity island named the locus of enterocyte effacement (LEE). The products of LEE are a type III secretion system, intimin and its translocated intimin receptor, and other secreted proteins. The secretion system is a molecular syringe for which secreted proteins are transferred into host cell cytoplasm. Intimin is encoded by eae gene that presents heterogeneity in their 3’ end, involved in binding to the enterocytes.
This study was the first in Iraq aimed to isolate and confirmed E. coli O157:H7serotypes in the faecal samples of calves located place around Baghdad province and to determine the eaeA virulence genes in these strains by PCR technique.
Materials and Methods
Bacteriological Examination
Three hundred fifty calves aged between (up to one year), from both sexes found in field at different places in Baghdad city, for six months (November 2014 to April 2015). All methods of culturing, Gram stain and biochemical test were done according to (Marky et al., 2014). Samples were cultured on MacConkey agar and Eosin Methylene blue agar and incubated aerobically at 37°C for 24- 48 hours; the growing colonies were examined by naked eye concerning their shape, size and color. Then bacterial cells were stained by gram stain. IMViC” tests (Indole, Methyl Red, Voges-Proskauer and Citrate) and Triple sugar iron medium. Culturing on two specific media, Cefixime Tellurite - Sorbitol MacConkey agar (CT-SMAC)[ LABM™ (England) ] and CHROM agar O157 [The Pioneer of Chromogenic Media/Paris] according to (Chow et al., 2006) and confirmation by using Latex agglutination Test for E. coli O157:H7. This test was used for serotyping of E. coli O157:H7 by using commercial kit (Wellcolex E. coli O157:H7, Remel) to detect both the somatic antigen O157 and the flagellar antigen H7 according to the manufacturer company.
PCR Assay for Detection of eaeA gene in Isolated E. coli O157:H7.
DNA extraction: Genomic DNA of E. coli O157:H7 isolate was extracted by using (Presto™ Mini g DNA Bacteria Kit Geneaid. USA) according to manufacture procedure.
Oligonucleotide primer: The oligonucleotide primers for eaeA gene were:
F -5’ GAC CCG GCA CAA GCA TAA GC -3’
and
R -5’ CCA CCT GCA GCA ACA AGA GG -3’
The product size was 384 pb were designed by (Paton and Paton, 1998). The purity and concentration of extracted DNA was measured using Nanodrop spectrophotometer (NuDrops)™ [ActGene(USA)] .
Gel electrophoresis for check extracted DNA: It was very important step to complete PCR assay, which was used to check the extracted DNA by loading the eluted DNA by agarose gel electrophoresis.
Preparation of PCR Master Mix: All required reagents were thawed completely and put them on ice, and reagent was mixed well by inversion and spins them down prior to pipetting. PCR master mix reaction was prepared by using GoTaq® Green Master Mix from Promega, USA.
The PCR tubes containing an amplification mixture were transferred to thermal-cycler and started the program for amplification as shown in the Table 1.
Table 1: PCR program for detection (eaeA) gene
|
No. of cycles |
Time |
Temperature (°C) |
Step |
|
1 |
5 min. |
94 |
Initial denaturation |
|
35 |
1min. |
94 |
Denaturation |
|
30 sec. |
57 |
Annealing |
|
|
1 min. |
72 |
Extension |
|
|
1 |
10min |
72 |
Final extension |
|
4 |
Hold |
PCR Product Analysis (Agarose Gel Electrophoresis): It is a very important step to complete PCR assay, which was used to analyse the PCR product by agarose gel electrophoresis, Finally PCR products (bands) were visualized using a UV transilluminator[ Cleaver Scientific (U.K.)] and photographed by using digital camera.
Ethically Approved
This study was approved by the ethical and research committee of Veterinary Medicine of College, University of Baghdad, Ministry of High Education and Scientific Research.
Results
E. coli O157:H7 Isolation
Different morphological shape and colour of E. coli colonies were appeared on different media. The colonies revealed red /pink colour On MacConkey agar and metallic sheen on Eosin Methylene Blue. The Gram stain of suspected E. coli colonies revealed, negative non spore forming rod. The isolated bacteria gave different reaction in biochemical tests. It gave negative for Voges–proskuar, simmon citrate and positive for indole and motility tests. The Triple sugar Iron test (TSI) showed Yellow with/without gas production. The isolated colonies of E. coli appeared small, circular and colourless with smoky centre (1-2) mm in diameter on SMA-CT. On Chrome agar the colonies of E. coli O157 showed mauve colour.
Serotyping Test (Wellcolex E. coli O157:H7, Remel)
Escherichia coli colonies from (SMA-CT) were tested for identification of both O157:H7 antigens by Wellcolex E. coli O157:H7, Remel. The isolates that gave a positive reaction for the O157 antigen were sub-cultured overnight on blood agar for the detection of flagellar antigen (H7). Red colour agglutination indicated a positive result for (O antigen) in comparison to clear red colour of the control and the blue colour agglutination indicated positive result for (H antigen) in comparison to clear blue colour of the control.
Prevalence of E. coli O157:H7 in Calves
E. coli was isolated at a high percentage from samples of Diarrheic and non-diarrheic calves, E. coli O157:H7 appeared in 4 isolates (11.42%) from diarrheic calves, and 28 isolates of E. coli O157:H7 were isolated from non-diarrheic calves (Table 2).
Table 2: Number and rate of infection of E. coli O157:H7 in diarrheic and healthy calves
|
No. of E. coli O157:H7 |
No. of E .coli isolates |
No. of samples |
Animals |
|
4(11.42%) |
32(91.42%) |
35 |
Diarrheic calves |
|
28(8.88%) |
306(97.14%) |
315 |
Non-diarrheic calves |
|
32(9.14%) |
338(96.57) |
350 |
Total |
E. coli O157:H7 Confirmation by PCR
The confirmation process of the E. coli O157:H7 isolates recovered from fecal samples of calves to detect the presence of specific virulence trait eaeA gene by PCR assay, all four isolates from diarrheic calves were possess eaeA gene (100%) and 13 (46.42%) of isolates from non-diarrheic calves were positive for eaeA gene. The study revealed that 17(53.12%) from total isolates gave positive results with eaeA primers equal to target product size(384bp) (Table 3 and Figure 1).
Table 3: E. coli O157:H7 and % of virulence factor eaeA
|
No. of eaeA positive |
No. of E. coli O157:H7 |
|
4(100%) |
4(12.5%) |
|
13(46.42%) |
28(8.80%) |
|
17(53.12%) |
32(9.14%) |
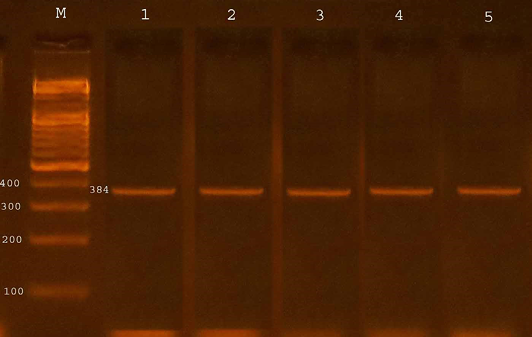
Figure 1: Agarose gel electrophoresis showed amplification of 384 bp fragments of eaeA genes of E. coli O157:H7
Lane M shows PCR marker
Discussion
This is the first study which describes the detection and frequency of major virulence genes of STEC isolated from cattle in Baghdad, Iraq. Study revealed 4 isolates of E. coli O157:H7 from 35 fecal samples at a percent (11.42%) in diarrheic calves and all these isolates possessed eaeA gene. Non diarrheic calves showed 28(8.88%) positive samples and 13(46.42%) possessed eaeA gene.
Escherichia coli O157:H7 are generally recognized by culturing on different media, on sorbitol MacConkey agar supplemented with cefixime and potassium tellurite (CT-SMAC), these results were compatible with Garcia et al. (2010) were they found that typical E. coli O157:H7 appeared as colourless colonies and do not fermented sorbitol on SMAC agar while most non-O157 strains ferment sorbitol and appear as pink colour colonies on SMAC agar. Another group of researcher, Tahamtan et al. (2011) use sorbitol-MacConkey agar plate supplemented with potassium tellurite (2.5 mg/L) and variant cifixime (0.05 mg/L). The E. coli O157:H7 on SMAC agar O157 colonies appear clear due to their inability to ferment sorbitol unlike other E. coli serotypes. Laegreid et al. (1999) also used sorbitol-MacConkey agar SMAC plates containing cifixime (0.5 mg), and potassium tellurite (2.5 mg) for isolation of E. coli O157:H7 from calves, After 18 hour incubation at 37°C the sorbitol negative colonies appear colourless.
Our results showed that Chrome agar aids in diagnosis of E. coli O157:H7, it utilizes one of chromogenic substrates which produce mauve colour colonies, while non- E. coli O157:H7 organism may utilize chromogenic substrates resulting in blue to blue green colour colonies, our results are in agreement with Tavakoli et al. (2008) who recorded that the use of chrome agar allowed presumptive identification of E. coli O157:H7 from the primary isolation plate and differentiation from other organisms. A similar study by Yousif and Al-Taii (2014) reported that the Chrom agar is useful for diagnosis of Escherichia coli 0157:H7.
Latex agglutination test appeared a highly sensitive and specific for the diagnosis of E. coli O157:H7, this results in agreement with Yousif and Al-Taii (2014), Al-Dawmy and Yousif (2013) and Karmali et al. (1999) who used latex agglutination test for serotyping of E. coli O157:H7. And describe it as a rapid, reliable, easy to perform and interpret, and it should allow testing for VT to become more widely performed.
The percentage of E. coli O157 isolation from calves were compatible with Omisakin et al. (2003) they reported the prevalence of carriage of E. coli O157 in faeces of cattle was 7.5% and with study of Alam and Zurek (2006) who found the prevalence of Escherichia coli O157:H7 in beef cattle faeces was (9.2%) Another study conducted by Kang et al. (2004) was compatible with our study as they found the prevalence of E. coli O157 in diarrheic calves at percentage 9.8% and with Kuyucuoglu et al. (2011) as they estimate the prevalence of E. coli O157:H7 in diarrheic calves at percentage (10.6). Whereas Blanco et al. (1993) found that the prevalence of Eschrechia coli O157:H7 in the faeces of dairy calves and feedlot cattle is low (0.3 to 2.2%) in the United States, the United Kingdom, Germany, and Spain. While Mechie et al. (1997) recorded the prevalence of Escherichia coli O157:H7 in calves a high percentage (56%) in England.
The prevalence of E. coli O157:H7 in the current study was higher than that reported by El-Shehedi et al. (2013) in AL-Qalyoubia Governorate in Egypt in diarrhoeic calves at level 6.97%.
In non-diarrheic calves, the results showed that the prevalence of E. coli O157:H7(8.88%) was higher than the percentage recorded by Kuyucuoglu et al. (2011), as they found 2.6% of healthy calves infected with E. coli O157:H7.
The occurrence of E .coli O157:H7 were also detected in different regions of Turkey. For instances, E. coli O157 was found in 14 individuals among 330 cattle slaughtered in five different abattoir in Istanbul (Yilmaz et al., 2002) and E. coli O157:H7 were isolated in 4 individuals among 312 cattle in the eastern region of Turkey (Aslantas et al., 2006). In another study, the rate of E. coli O157:H7 infection was found to be 13.6% (Cabalar et al., 2001), this point was very important because turkey was a neighbouring country to Iraq.
The results showed that eaeA gene found in a percentage (53.12%) in isolates of E. coli O157:H7 from diarrheic and non-diarrheic calves. This results agreed with Galland et al. (2001) who found that 26 from 57 Escherichia coli O157:H7 beef cattle feedlots in southwest Kansas were eaeA gene positive. But other researcher record a high percentage (100%), Schouten et al. (2004) found all Escherichia coli O157 isolates on Dutch dairy farms show positive for eaeA gene, Synge et al. (2003) recorded that all of the VTEC O157 tested were eaeA positive from beef suckler cows in Scotland and Alam and Zurek (2006) showed that all tested isolates of Escherichia coli O157:H7 in beef cattle were positive for eaeA (Intimin) gene.
Gene eaeA (Intimin) which was a necessary gene for attaching and effacing activity (Kaper et al., 1998). Many investigators have underlined the strong association between carrying eaeA and the capacity of STEC to cause severe human disease, especially HUS (Oswald et al., 2000). In the present study, this important virulence gene was detected in 100% of STEC O157:H7 in diarrheic calves and (46.42%) of non-diarrheic calves isolates. A similar prevalence of the intimin (eaeA) gene has also been found in other studies (Blanco et al., 2001; Blanco et al., 2004).
Epidemiological studies on EHEC in cattle are very necessary to develop control measures in order to reduce the risk of transmission from cattle to humans. Since isolation procedures are laborious and time-consuming and because of the lack of biochemical features distinguishing most EHEC strains from nonpathogenic E. coli, PCR approaches based on the detection of EHEC-associated genetic markers have been developed (Bibbal et al., 2014).
In conclusion, this study revealed the importance of E. coli O157:H7 serotype calves, which represent as a reservoir for strains that transmitted the disease to human. E. coli O157:H7 virulence gene eaeA (intimine) was detected in faeces samples collected from calves using PCR. Therefore, we believe that the isolation of E. coli O157:H7 serotypes in Iraq will be beneficial to the many researchers in this field and for investigation of future epidemiological study.
Acknowledgments
This work was supported by College of Veterinary Medicine, Department of Internal and Preventive Veterinary Medicine, University of Baghdad, Iraq.
Author’s cONTRIBUTION
Both authors contributed equally in all details of this manuscript.
Conflict of Interest
Authors declares no conflict of interest.
References






