The Journal of Advances in Parasitology
Research Article
Simultaneous Diagnosis and Species Identification of Microsporidial Infection in Human Stool Samples using Real-Time Polymerase Chain Reaction
Eman Dorry El-Kerdany1, Shwikar Mohamed Ahmed2, Maha Reda Gaafar1*, Radwa Galal Diab1, Eman Attia El-Morsy1
1Department of Medical Parasitology; 2Department of Medical Microbiology and Immunology, Faculty of Medicine, Alexandria University, Egypt.
Abstract | This study evaluated the utility of SYBR Green real-time PCR for simultaneous detection and differentiation of microsporidial infections in hundred stool samples of immunosuppressed patients in comparison to the modified trichrome stain, and in relation to the age, sex, and different causes of immunosuppression of the patients. DNA was extracted using ISOLATE Faecal DNA Kit and amplification was performed in a light cycler using a SensiFASTTMSYBRHi-ROX PCR kit using MsRTf1 and MsRTr1 primers. Of 100 stool samples routinely analysed for microsporidian spores, 49 were positive by microscopy. By measuring the spore size using micrometre, determination of the species of the positive cases was 17 Encephalitozoon intestinalis, 15 Enterocytozoon bieneusi, 11 Encephalitozoon hellem, and six Encephalitozoon cuniculi based on the reference spores’ size of each species. By SYBR Green real-time PCR, 55 stool samples were positive for microspoidial DNA upon determination of their specific melting curves, comprising 16 Encephalitozoon cuniculi, 14 Encephalitozoon hellem, 13 Encephalitozoon intestinalis, 10 Enterocytozoon bieneusi, and two unspecified species with melting temperature of 84.2018 and 84.903˚C. There was a positive agreement reaching 84 % between both techniques as regard the number of positive cases. Moreover, real-time PCR was superior over microscopy in species identification with statistically significant difference between both methods. However, there was no statistically significant difference between patients’ age, sex and causes of immunosuppression with different Microsporidia species detected by real-time PCR. Thus, SYBR Green real-time PCR can be considered a fast and reliable method for detection and identification of Microsporidia species.
Keywords | Microsporidia species, Modified trichrome stain, Spore’ size, SYBR Green real-time PCR, Melting curve
Editor | Muhammad Imran Rashid, Department of Parasitology, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | February 02, 2016; Accepted | July 04, 2016; Published | August 14, 2016
*Correspondence | Prof. Dr. Maha Reda Gaafar, Department of Medical Parasitology, Faculty of Medicine, Alexandria University, Egypt; Email: [email protected]
Citation | El-Kerdany ED, Ahmed SM, Gaafar MR, Diab RG, El-Morsy EA (2016). Simultaneous diagnosis and species identification of microsporidial infection in human stool samples using real-time Polymerase Chain Reaction. J. Adv. Parasitol. 3(4): 104-116.
DOI | Http://dx.doi.org/10.14737/journal.jap/2016/3.4.104.116
ISSN | 2311-4096
Copyright © 2016 El-Kerdany et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Microsporidia are eukaryotic, spore forming intracellular parasites belonging to the phylum Microspora that invade vertebrates and invertebrates. They are widely distributed in nature with over 1200 characterized species (Franzen and Muller, 1999). Microsporidiosis is primarily found in immunocompromised patients; especially those infected with Human Immunodeficiency Virus (HIV) or have undergone organ transplantation. Some species, however, have also been known to parasitize those with competent immune systems. Beyond humans, Microsporidia are important parasites in fisheries, veterinary medicine and pest management. Microsporidia are considered causal, accidental or opportunistic agents in humans (Franzen and Muller, 1999; Fournier et al., 2000; Gumbo et al., 2000).
The worldwide epidemiology of human microsporidiosis is poorly understood, and recent data suggest that the incidence of these pathogens is much higher than previously reported and may represent a neglected infectious agent in immunocompetent individuals (Luna et al., 1995; Ojuromi et al., 2012).
Seven genera (Enterocytozoon bieneusi (E. bieneusi), Encephalitozoon species {Encephalitozoon intestinalis (E. intestinalis), Encephalitozoon cuniculi (E. cuniculi), Encephalitozoon hellem (E. hellem)}, Pleistophora, Trachipleistophora, Nosema, Vittaforma, and unclassified Microsporidia which is referred to by the collective term Microsporidium) have been confirmed to cause human infection. The spectrum of the disease includes gastrointestinal, renal, pulmonary, nasal, ocular, muscular, cerebral, and systemic infections (Franzen and Muller, 1999; Tosoni et al., 2002; Chabchoub et al., 2009).
To date, microsporidial infections due to E. bieneusi and E. intestinalis are common and they have been identified as important agents for chronic diarrhea and wasting syndrome (Sak et al., 2010). These infections are found most commonly in immunocompromised patients; however, E. bieneusi has also been detected in the stool samples of immunocompetent individuals (Gumbo et al., 2000).
The prevalence of microsporidiosis is likely underestimated due to the labour-intensive, insensitive, and nonspecific laboratory staining methods used for the diagnosis of this disease (Luna et al., 1995; Franzen and Muller, 1999). Although the organisms can be identified in routine histological preparations, they don’t take the stain properly (Garcia et al., 1994). Moreover, routine staining of clinical specimens can be performed using a Weber’s modified trichrome stain (MTS) which does not differentiate spores at species level. The spores of these parasites can be identified as small (1-4 µm) oval bodies that exhibit fluorescence when stained with calcofluor white or other fluorescent brighteners (Luna et al., 1995; Kumar et al., 2005; Garcia, 2007). Thus, while the staining procedures are common they require skilled and experienced technologists that contributes to under reporting of this infection.
In fact, transmission electron microscopy (TEM) was reported to be the best approach for the identification of Microsporidia to the genus level, by examining the morphological characteristics of the polar filaments. However, inspite of considering the electron microscopy as the gold standard for species identification, morphology alone will not always allow identification of Microsporidia up to the species level. As TEM is an expensive, time-consuming and unavailable for routine diagnosis at all laboratories, molecular methods are increasingly being developed and employed for diagnosis of these organisms (Kumar et al., 2005; Joseph et al., 2006; Chabchoub et al., 2009; Ghosh and Weiss, 2009; Van Lieshout and Verweij, 2010).
Traditional polymerase chain reaction (PCR) ameliorates the diagnosis of Microsporidia in some clinical laboratories. Many of the available assays have been developed for research purposes; however, they are not approved to be among routine testing. This is because of their tedious sample processing and DNA extraction methods, in addition to the false-positive results that are possibly caused by specimen contamination, and unexperienced technologists. Besides, false negative results that could occur because of minimum organism concentration below the lower limit of detection, inadequate specimen storage, or the presence of faecal inhibitors (Franzen and Muller, 1999; Orlandi and Lampel, 2000; Accoceberry et al., 2001; Garcia, 2002). A number of species-specific PCRs were previously used for the detection and differentiation of microsporidian spores in different clinical specimens. However, the growing need to cover the full spectrum of human parasites could represent significant financial and technical barriers for routine diagnosis and hence, real-time PCR technique has been improved for detection and differentiation of E. bieneusi and Encephalitozoon species (Polley et al., 2011). Moreover, we are indeed to identify such resistant Microsporidia species (E. bieneusi) to be followed by trials for discovery of effective antimicrobial agents.
The qualitative real-time PCR with SYBR Green dye and DNA melting curve analysis is a technique capable of identifying multiple species in different samples containing DNA. It involves the use of SYBR Green fluorescent dye which binds to double stranded DNA and fluoresce (Arya et al., 2005). The shape and the position of the DNA melting curves can be used to differentiate between amplification products which can be separated by less than 2oC in melting temperature (Ririe et al., 1997).
The aim of the present study was to evaluate the utility of SYBR Green real-time PCR for the simultaneous detection and differentiation of enteric microsporidial infections in stool samples of immunosuppressed patients in comparison to the conventional MTS, and in relation to the age, sex, and different causes of immunosuppression of the patients.
Material and Methods
One hundred stool samples were randomly collected from immuno-compromised patients (50 males and 50 females) complaining of GIT problems, mainly diarrhea from May to October 2014, with no predilection for patients’ age or sex. Fifty-four patients were suffering from haematological malignancies (lymphoma& leukaemia), 23 with chronic renal failure, 14 were receiving immunosuppressive drugs as steroid and chemotherapeutic agents, and 9 patients had other causes like diabetes mellitus, idiopathic immunodeficiency and aplastic anaemia. The samples were collected from Alexandria University Hospital and Fever Hospital (Alexandria, Egypt), and written informed consents were obtained from the patients. Stool specimens were tested at the Diagnostic Laboratory of the Parasitology Department, Faculty of Medicine, Alexandria University, Egypt. Each sample was divided into two parts; the first part was preserved into 2.5% potassium dichromate, and stored at 4˚C for further microscopic examination and staining (Liguory et al., 2000; Williams et al., 2010). The second part was freshly frozen at -70˚C to be used for the molecular study (Salih et al., 2012). This study was approved by the Ethics Committee of Alexandria University.
Parasitological Study
The stool samples preserved in potassium dichromate were examined by direct saline smear (Garcia, 2007; Zaglool et al., 2011), iodine stained smear (Franzen and Muller, 1999; Garcia, 2007; Zaglool et al., 2011), and ether sedimentation technique (Muller et al., 1999), to detect other parasites as a cause of diarrhea.
All faecal samples were filtered by Lumb’s filtration technique (Lumb et al., 1993), then stained by MTS (Weber et al., 1992), in order to detect the microsporidial spores clearly without faecal debris. In addition, modified Ziehl Neelsen stain (MZN) was performed for detection of coccidial infection (Garcia, 2007).
Spore Measurement
Spore measurement in the modified trichrome stained smears was done using the micrometre in order to obtain species identification based on the reference size of the spores of each species which is 1-1.6 ×0.6-0.9 µm for E. bieneusi, 1.5-2×1-1.2 µm for E. intestinalis, 2.5-3.2×1.2-1.6 µm for E. cuniculi, and 2- 2.5 ×1- 1.5 µm for E. hellem (Petry, 2000). Reading was carried out in six different spores in each examined slide, and then average size of the spores was calculated (Fleck and Moody, 1993; Monica, 1998; Moss et al., 1999).
Molecular Study
The frozen stool samples were used for the diagnosis and identification of different species of Microsporidia. This was achieved by qualitative real-time PCR with SYBR Green dye and DNA melting curve analysis (Ririe et al., 1997; Arya et al., 2005; Polley et al., 2011). DNA was extracted from stool samples using ISOLATE Faecal DNA Kit (Bioline Co. UK) according to the manufacturers’ instructions. In brief, faecal samples were added directly to a bashing beads lysis tube and they were rapidly lysed by bead beating in a vortex, without the use of organic denaturants or proteinases. The DNA was then bound, isolated and purified using spin columns. The resulting DNA extracts were stored at -70°C until PCR assessment. Primers MsRTf1 (5’-CAGGTTGATTCTGCCTGACG-3’) and MsRTr1 (5’-CCATCTCTCAGGCTCCCTCT-3’) (Metabion International AG. Germany) were used for the amplification of E.intestinalis, E. hellem, E. cuniculi, and E.bieneusi SSU-rRNA sequences. Amplification was performed in a light cycler (Applied Biosystem, One Step, California, USA) using a SensiFASTTMSYBRHi-ROX PCR kit (Bioline Co. UK). In short, forward and reverse primers (10 pmol each) were used in 20 µl reactions containing 3 µl of the DNA extract. The reaction was performed with 35 amplification cycles of denaturation at 95˚C for 10 s, annealing at 60˚C for 10 s, and primer extension at 72˚C for 20 s. This was followed by melting point analysis of the PCR product duplex consisting of 95˚C for 10 s, followed by cooling to 70˚C for 20 s. The temperature was raised to 95˚C at a rate of 0.2˚C/s with continuous fluorescence acquisition (Ririe et al., 1997; Arya et al., 2005; Polley et al., 2011).
Upon observing the specific melting curves, stool samples were considered to be microsporidial DNA positive (Polley et al., 2011). DNA extracts of Microsporidia produce melting curves with melting temperatures of 81.4 - 81.9˚C for E. bieneusi, 82.7 - 83.5 ˚C for E. intestinalis, 83.6 - 84˚C for E. cuniculi, and 82 - 82.4 ˚C for E. hellem.
Statistical Analysis of the Data
Data were fed to the computer and analysed using IBM SPSS software package version 20.0. Qualitative data were described using number and percent. Quantitative data were described using mean and standard deviation. Comparison between different groups regarding categorical variables was tested using Chi-square test, and when more than 20% of the cells have expected count less than 5, correction for Chi-square was conducted using Monte Carlo correction. Agreement between the different predictive with the outcome was expressed in sensitivity, specificity, positive predictive value and negative predictive value. Agreement for PCR with microscopy was done using kappa test. A non-parametric test (Kruskal Wallis test) was used to compare between different groups. Significance of the obtained results was judged at the 5% level (Kotz et al., 2006; Kirkpatrick and Feeney, 2013).
Results
On the basis of the results of the reference methods, out of the 100 stool samples collected and examined microscopically, 49 were positive for Microsporidia spores (Figure 1), and six samples contained Cryptosporidia oocysts. Moreover, Blastocystis hominis cysts, Giardia lamblia cysts and Entamoeba coli cysts were observed in the samples. It has to be noted that Microsporidia spores, Blastocystis hominis cysts, and Cryptosporidia oocysts were only detected following staining techniques.
When the micrometre was used to measure the dimensions of spores in modified trichrome stained smears, 17 cases (34.7%) were E. intestinalis with size between 1.5-2 × 1-1.2 µm. Fifteen cases (30.6%) were coinciding with E. bieneusi whose size ranged from 1.3-1.6 ×0.7-0.9 µm. Eleven cases (22.4%) were considered E. hellem with a size 2-2.3 × 0.9-1.5 µm, and eventually six cases (12.2 %) were consistent with E. cuniculi with a size 2.5-3 × 1.2-1.4 µm. (Table 1) As this technique is a subjective measure, it could not identify the Microsporidia species definitely. Thus, we considered this technique as a preliminary step in species identification.
Table 1: The average spore size in µm of 49 positive samples measured by ordinary micrometre
|
E. bieneusi |
E. intestinalis |
E. cuniculi |
E. hellem |
|
1.55x 0.7 |
1.7x 1.1 |
3 x 1.4 |
2.04 x 1.19 |
|
1.6 x 0.8 |
1.85 x1 |
2.5 x 1.2 |
2.1x1.3 |
|
1.45 x 0.7 |
1.7 x 0.9 |
2.95 x 1.15 |
2.1 x 1.45 |
|
1.6 x0.8 |
2 x 0.9 |
2.9 x1.35 |
1.91 x 1 |
|
1.59 x 0.7 |
1.4 x 1.2 |
2.5 x 1.19 |
2 x 0.9 |
|
1.55 x0.8 |
2 x 0.9 |
2.9 x 1.2 |
2 x 1 |
|
1.58x 0.9 |
2 x1.19 |
1.99 x 0.97 |
|
|
1.3x0.9 |
1.5 x1.2 |
2.1 x1 |
|
|
1.6 x 0.9 |
1.58 x 0.95 |
2.3 x 0.99 |
|
|
1.6 x 0.89 |
1.53 x 1 |
2.25 x 0.9 |
|
|
1.6 x 0.8 |
1.7 x 0.97 |
2.1 x 0.9 |
|
|
1.59 x 0.7 |
2 x 1.2 |
||
|
1.58 x 0.8 |
2 x 1 |
||
|
1.5 x 0.9 |
1.6 x 1.1 |
||
|
1.5 x 0.8 |
1.9 x 1 |
||
|
1.7 x 1.1 |
|||
|
1.96 x 0.6 |
In the present work, total DNA was extracted successfully from the 100 stool samples using the ISOLATE Faecal DNA Kit, which provides sufficient amounts of pure DNA for PCR testing. SYBR Green real-time PCR assay using the primer pair in this study resulted in efficient amplification of Microsporidia DNA fragments of 55 samples (55%). Amplification failed in the remaining 45 DNA extracts, thus they were considered negative.

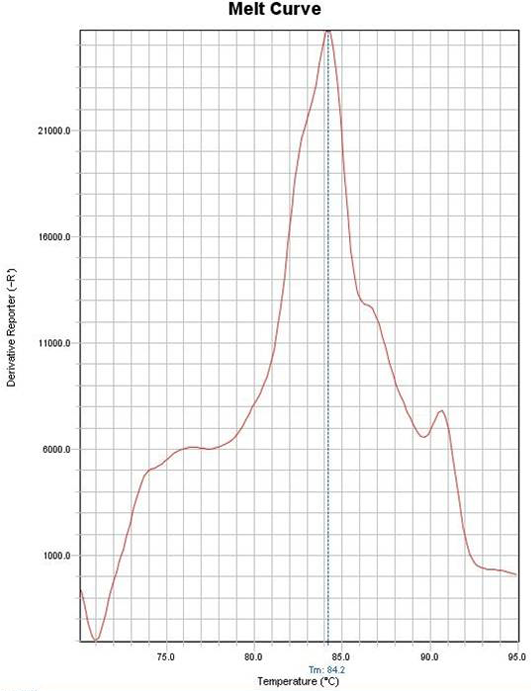
Figure 2: Example of melting curve of one case diagnosed as Encephalitozoon cuniculi with melting temperature of 84˚C
A total of 55 Microsporidia–positive samples were identified to the species level according to their reference melting temperatures. Out of 55, 16 cases (29.1%) were consistent with E. cuniculi whose melting temperature ranged from 83.6˚C - 84.09˚C (Figure 2), 14 samples constituting 25.5% of the positive cases were for E. hellem with melting temperature between 82.1˚C - 82.4˚C. E. intestinalis was detected in 13 cases (23.6%) and the melting temperature ranged from 82.6˚C - 83.39˚C. Ten samples (18.2%) were for E. bieneusi with a melting temperature range from 81.33˚C - 81.9˚C. Finally, two samples (3.6%) were considered as unspecified species, as they showed single bands with melting temperature of 84.2018 and 84.903˚C (Figure 3) (Table 2). Negative control samples were set and their melting temperature curves were plotted in Figure 4. It has to be noted that both unspecified species by PCR were previously identified to be E. bieneusi and E. intestinalis respectively depending on their spores’ size (1.5 × 0.8µm &1.96 × 0.6µm).
Table 2: The melting temperature in oC of the 55 positive stool samples for Microsporidia detected by SYBR Green real-time PCR
|
E. bieneusi |
E. intestinalis |
E. cuniculi |
E. hellem |
Unspecified species |
|
81.33 |
83.2007 |
84.0967 |
82.3169 |
84.2018 |
|
81.42 |
82.6075 |
83.8976 |
82.3169 |
84.903 |
|
81.8193 |
82.615 |
83.6042 |
82.209 |
|
|
81.719 |
82.6079 |
83.7038 |
82.4082 |
|
|
81.91 |
82.7072 |
83.6039 |
82.4082 |
|
|
81.918 |
82.615 |
83.9029 |
82.1093 |
|
|
81.8019 |
82.7072 |
83.7036 |
82.1093 |
|
|
81.8019 |
83.1084 |
83.6039 |
82.4082 |
|
|
81.9146 |
83.2081 |
83.8032 |
82.1093 |
|
|
81.9146 |
82.909 |
83.9029 |
82.4164 |
|
|
82.7096 |
83.9029 |
82.217 |
||
|
82.6099 |
83.9029 |
82.418 |
||
|
83.2956 |
83.8032 |
82.114 |
||
|
83.909 |
82.117 |
|||
|
83.7935 |
||||
|
84.008 |
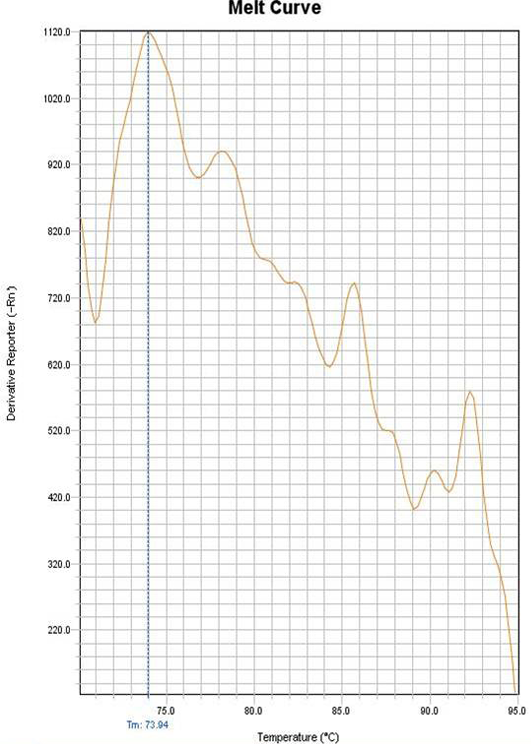
Figure 4: Melting curve of the negative control
Table 3: The number of Microsporidia positive and negative stool samples by both studied diagnostic techniques
|
SYBR Green real-time PCR |
Microscopy |
Total |
|
|
Total No. of +ve cases |
Total No. of -ve cases |
||
|
Total No. of +ve cases |
44 |
11 |
55 |
|
Total No. of -ve cases |
5 |
40 |
45 |
|
Total |
49 |
51 |
100 |
As regards the agreement of MTS method with SYBR Green real-time PCR in diagnosing microsporidial cases, we found that 44 of the positive cases for Microsporidia by MTS microscopy were also positively detected by SYBR Green PCR, thus, they were considered true positive. Meanwhile, 40 of the negative cases for Microsporidia by MTS microscopy were also reported negative with the SYBR Green PCR, and hence reported as true negative samples. On the other hand, five cases out of the 49 positive cases of the Microsporidia by microscopy were negative by SYBR Green PCR, thus they were considered false negative. However, 11 cases were positive by SYBR Green real-time PCR only and were considered false positive samples (Table 3). Thus, in light of the previous statements, and when microscopy was used as the reference standard, the sensitivity, the specificity, negative and positive predictive values of the of the SYBR Green real-time PCR were 89.8%, 78.43%, 80 % and 88.89% respectively with a degree of agreement that reached 84 % (Table 4). According to Chi-square Test, microscopic examination and SYBR Green real-time PCR showed no statistically significant difference in the number of the positive cases for Microsporidia in stool samples (Table 5).
Table 4: Agreement between the diagnostic procedures used for the detection of Microsporidia in the 100 stool samples
|
Relation between the results of Microscopy and SYBER Green real-time PCR |
Number of stool samples |
|
Agreement |
|
|
Positive / positive |
44 |
|
Negative / negative |
40 |
|
Disagreement |
|
|
Positive / negative |
5 |
|
Negative / positive |
11 |
Agreement %: 84%
Table 5: The number of positive and negative samples of Microsporidia by microscopic examination and SYBR Green real-time PCR
|
Microscopic examination |
SYBR Green real-time PCR |
|||
|
Microsporidia |
Number |
Percent |
Number |
Percent |
|
+ve |
49 |
49 % |
55 |
55 % |
|
-ve |
51 |
51 % |
45 |
45 % |
|
P |
0.396 |
|||
P: value of Chi-square test for comparison between microscopic examination and SYBR Green PCR; *Statistically significant P ≤ 0.05
When considering the true positive cases of both diagnostic techniques (44 cases) and after exclusion of the two unspecified species by SYBR Green real-time PCR, the species identified for 42 cases by both ordinary micrometre and SYBR Green real-time PCR were compared. When PCR was used as the gold standard for species identification, results were as follows; micrometre based method showed positive agreement with real time PCR method in 20 cases, where typical species were identified in both techniques using the reference spore dimensions and the melting temperature curves respectively. However, ordinary micrometre method failed to agree with typical species detected by real time PCR in 22 cases with statistically significant difference in the capability of both diagnostic techniques in species identification (Table 6).
In this study, the samples were randomly collected; however, statistical analysis of the clinical data of the patients shared in determining some factors that could affect the type of Microsporidia infection acquired. Regarding the age of the patients, the relation between the type of Microsporidia species detected by real-time PCR and the patient’s age were shown in Table 7. According to Chi-square for Kruskal Wallis test, there was no statistically significant difference between the various microsporidial species and the patients’ age. On the other hand, on studying the sex as demographic factor that may affect the species of microsporidial infection, it was found that out of the 55 Microsporidia positive cases proved by PCR; 30 cases were females and 25 were males. The relation between the type of Microsporidia species detected by real-time PCR and the patient’s sex were shown in Table 8. According to Chi-square and Monte Carlo tests, there was no statistically significant difference between patients’ sex and different Microsporidia species. Besides, the relation between the Microsporidia species detected by real-time PCR in the immunosuppressed patient’s stool and the cause of immunosuppression was analysed and represented in Table 9, which showed that there was no statistically significant difference between causes of immunosuppression and species of Microsporidia.
Table 6: The percentage of agreement between the ordinary micrometre and SYBR Green-real time PCR in detection of Microsporidia species in 42 cases
|
SYBR green real time PCR |
χ2 |
MCP |
||||||||
|
E .bieneusi (n = 8) |
E. intestinalis (n=10) |
E. cuniculi (n = 13) |
E. hellem (n=11) |
|||||||
|
Ordinary Micrometre |
No. |
% |
No. |
% |
No. |
% |
No. |
% |
||
|
E. bieneusi |
3 |
37.5 |
0 |
0.00 |
0 |
0.00 |
6 |
54.5 |
28.876* |
<0.001* |
|
E .intestinalis |
3 |
37.5 |
9 |
90.0 |
4 |
30.8 |
0 |
0.00 |
||
|
E. cuniculi |
0 |
0.00 |
1 |
10.0 |
4 |
30.8 |
1 |
9.1 |
||
|
E. hellem |
2 |
25.0 |
0 |
0.00 |
5 |
38.5 |
4 |
36.4 |
||
|
Kappa |
0.307* |
|||||||||
|
p |
<0.001* |
|||||||||
|
Agreement |
Fair agreement |
|||||||||
χ2: value for Chi-square test; MC: Monte Carlo; Κ: kappa test; *: Statistically significant at p ≤ 0.05
Table 7: Distribution of 55 Microsporidia positive cases according to different age groups, and the relation between the different Microsporidia species detected by SYBR Green real-time PCR and patients’ age
|
Age groups |
Species of Microsporidia |
||||
|
E. bieneusi (n=10) |
E. intestinalis (n=13) |
E. cuniculi (n=16) |
E. hellem (n=14) |
Unspecified species (n=2) |
|
|
0 -10 years |
7 |
5 |
12 |
7 |
1 |
|
11- 20 years |
1 |
0 |
1 |
3 |
0 |
|
21 -30 years |
1 |
0 |
0 |
1 |
0 |
|
31 -40 years |
0 |
4 |
1 |
1 |
1 |
|
41- 50 years |
1 |
0 |
0 |
1 |
0 |
|
51- 60 years |
0 |
1 |
1 |
0 |
0 |
|
61 -70 years |
0 |
2 |
0 |
1 |
0 |
|
71 -80 years |
0 |
1 |
0 |
0 |
0 |
|
81 -90 years |
0 |
0 |
1 |
0 |
0 |
|
Age Mean ± SD |
9.31 ± 13.78 |
31.96 ± 26.88 |
14.57 ± 22.75 |
16.46 ± 18.37 |
17.50 ± 20.51 |
|
KWχ2 |
5.945 |
||||
|
p |
0.203 |
||||
KWχ2: Chi square for Kruskal Wallis test; *: Statistically significant at p ≤ 0.05
Table 8: The relation between the Microsporidia species detected by SYBR Green real-time PCR and patients’ sex
|
Species of Microsporidia |
||||||||||
|
E. bieneusi (n = 10) |
E. intestinalis (n = 13) |
E. cuniculi (n = 16) |
E. hellem (n = 14) |
Unsecified species (n = 2) |
||||||
|
Sex |
No. |
% |
No. |
% |
No. |
% |
No. |
% |
No. |
% |
|
Male |
7 |
70.0 |
6 |
46.2 |
6 |
37.5 |
6 |
42.9 |
0 |
0.0 |
|
Female |
3 |
30.0 |
7 |
53.8 |
10 |
62.5 |
8 |
57.1 |
2 |
100.0 |
|
χ2 |
4.131 |
|||||||||
|
MCp |
0.389 |
|||||||||
χ2: value for Chi-square test; MC: Monte Carlo; *: Statistically significant at p ≤ 0.05
Table 9: The relation between the Microsporidia species detected by SYBR Green real-time PCR with the causes of immunosuppression
|
Species of Microsporidia |
||||||||||
|
E. bieneusi |
E. intestinalis |
E. cuniculi |
E. hellem |
Unspecified species (n = 2) |
||||||
|
Disease |
No. |
% |
No. |
% |
No. |
% |
No. |
% |
No. |
% |
|
Aplastic anemia |
0 |
0.00 |
0 |
0.0 |
2 |
12.5 |
0 |
0.0 |
0 |
0.0 |
|
Autoimmune Hemolytic anemia |
0 |
0.0 |
1 |
7.7 |
0 |
0.0 |
0 |
0.0 |
0 |
0.0 |
|
Chronic steroid therapy |
2 |
20 |
2 |
15.4 |
2 |
12.5 |
2 |
14.3 |
0 |
0.0 |
|
Diabetes mellitus |
0 |
0.0 |
0 |
0.0 |
0 |
0.0 |
1 |
7.1 |
0 |
0.0 |
|
Leukemia |
3 |
30.0 |
5 |
38.5 |
5 |
31.25 |
8 |
57.1 |
1 |
50.0 |
|
Lymphoma |
1 |
10.0 |
3 |
23.1 |
2 |
12.5 |
1 |
7.1 |
1 |
50.0 |
|
Renal failure |
4 |
40.0 |
2 |
15.4 |
5 |
31.25 |
2 |
14.3 |
0 |
0.0 |
|
χ2 |
25.144 |
|||||||||
|
MCp |
0.882 |
|||||||||
χ2: value for Chi square test; MC: Monte Carlo; *: Statistically significant at p ≤ 0.05
Therefore, in light of our results, E. bieneusi infection is more common in younger males with mean age of 9.31 years. E. cuniculi infection is more common in females with mean age of 14.57 years, while E. intestinalis and hellem affect nearly equally both males and females, with mean age of 31.9 and 16.4 years respectively, while in E. bieneusi, 70% of the infections were in males. Finally, infection with Encephalitozoon species is more common in haematological malignancies especially leukaemia, while infection with E. bieneusi infection is more common in patients with renal failure.
Thus, this comparative study has shown that the SYBR Green real-time PCR was superior over the microscopic techniques as it detected higher number of positive cases of Microsporidia than those detected by the staining techniques. Furthermore, it improved species determination by the melting temperature curves more than the species identified through measurement of the spores’ size by the micrometre in the stained smears.
Discussion
Diarrhea due to opportunistic parasites has gained a lot of interest due to the spread of immunosuppressive conditions. Enteric protozoa and sporozoa emerged as important causes of such fatal diarrhea. Among these organisms, Microsporidia have emerged as important parasites (Eckmann, 2002; Tuli et al., 2010). With increased awareness of the oportunistic infections and improved diagnostic techniques, Microsporidia has now been identified in a broader range of human populations that includes not only immunocompromised hosts, but also includes travellers, children, and the old people (Didier and Weiss, 2006).
Over years, detection of Microsporidia has been a challenge as they are usually missed by routine microscopy and staining. Furthermore, identification of Microsporidia to the species level can influence therapeutic intervention since infection with Encephalitozoon species can be successfully treated with albendazole, whereas those caused by E. bieneusi may be resistant to the same therapy (Curry and Smith, 1999; Garcia, 2007). In order to achieve this aim, several methods have been used, but many of these techniques are cumbersome, time consuming, and required specialized techniques as EM and conventional PCR (Moura et al., 1999; Tuli et al., 2008). Moreover, a number of species-specific PCRs were previously used for the detection and differentiation of microsporidian spores in clinical specimens as the PCR-restriction fragment length polymorphism (PCR-RFLP) (Katzwinkel et al., 1997), multiplex real-time PCR (Verweij et al., 2007), oligo-nucleotide microarrays (Wang et al., 2005), and PCR followed by differential hybridization (Notermans et al., 2005). These techniques are not only tedious but also are currently available in few laboratories. Therefore, rapid, easy and sensitive techniques are needed to provide an early diagnosis and species identification of this infection.
In the present work, detection of Microsporidia was carried out using conventional parasitological techniques by staining of stool samples with MTS followed by measuring the size of the detected spores by micrometre to identify the species of Microsporidia. A molecular study was performed on the same samples using SYBR Green real-time PCR to test its ability to detect and differentiate types of intestinal Microsporidia species by melting temperatures and in relation to age, sex and different causes of immunosuppression.
In the view of the seasonal variability of microsporidial infection and the high incidence of infection during hot seasons, samples were collected in the period from May to October 2014. This was coincided with El-Shazly et al. (2007), who proved that public water supplies in Egyptian governorates showed highest prevalence of polluting protozoa, including Microsporidia spores in hot weather which is considered ideal for viability and infectivity of protozoal cysts. Stool samples used for parasitological examination were preserved in 2.5% potassium dichromate till the time of testing as it conserves the morphological properties of different parasitic stages in stool. On the other hand, samples used for molecular study were frozen fresh at -70˚C to avoid destruction of the DNA present in the stool sample and to avoid interference of the preservative with the PCR testing. This was documented with many stool preservatives especially formaldehyde (Da Silva and Pieniazek, 2003).
In the present study, while it has obviously been demonstrated that microscopy is not completely efficient in diagnosing microsporidial infections, it is generally identified as the gold standard for the diagnosis of parasitic diseases, against which new tests are compared. It has the advantages of being inexpensive and available in all laboratories. It also could detect different parasites in the stool samples. This was agreed with Marshall et al. (1997) and Ndao (2009).
In the current study, Microsporidia spores were only detected following staining techniques. This could be attributed to the small size of the Microsporidia spores that requires proper staining to make evident of the spore’s wall and polar eminent (Weber et al., 1992; Petry, 2000; Lobo et al., 2014). Furthermore, the masking effect of stool debris in the examined sediment on the tiny organisms could contribute to under reporting of these infections. That’s why we implemented stool filtration using Lumb’s technique in order to remove excess debris and to ensure that spores will be evident in a clear background after staining (Lumb et al., 1993).
In order to fulfil our aim, and in light of reference data on dimensions of different types of spores, we classified our Microsporidia positive samples after measuring the detected spores. However, we considered this technique as a preliminary step and could not identify the Microsporidia species properly. This was in accordance with Corcoran et al. (1995) and Ombrouck et al. (1997), who reported that identification of Microsporidia species by the spores’ size in stained stool samples could be achieved, but, it is considered as a primary diagnosis and there is a need for further confirmation by other more accurate methods. Moreover, Franzen et al. (1995) stated that the spore size may allow differentiation between Enterocytozoon and Encephalitozoon species, but cannot differentiate between the individual members of Encephalitozoon species.
On the other hand, as regards the SYBR Green real-time PCR results, DNA was easily and rapidly extracted from the stool samples by the used ISOLATE Faecal kit without any contaminants and enzyme inhibitors as suggested by the manufacture. (Bioline Co. UK) Species-specific universal PCR primer was used with real-time PCR to amplify DNA of the four species; E. bieneusi, E. intestinalis, E. hellem and E. cuniculi, considering previous records that enlisted them the most common species causing human infection (Franzen and Muller, 1999; Wang et al., 2005). Then, melting-curve analysis was achieved to differentiate between species by observing specific curves, thus eliminating the extra step of gel analysis. It was previously recorded that the real-time PCR method is very sensitive and has been used in diagnosis of many pathogens (Yu et al., 2005; Burgher-MacLellan et al., 2009).
In the current study, molecular testing showed 55% positive stool samples for Microsporidia. When considering the microscopy as a reference standard technique, eleven cases were considered to be false positive and five cases were recorded as false negative. The presence of false positive cases could be explained by Ignatius et al. (1997) who stated that detection of spores in stool samples using ordinary light microscope required well trained expertise. Also, Fayer et al. (2003) and Verweij et al. (2007) reported that the lowest detection limit of light microscopy has been recorded to be between 104 and 106 microsporidial spores / g stool, whereas real time PCR could detect as few as one spore / g stool. In addition, Polley et al. (2011) suggested that microsporidial infections in the tissues may result in the presence of only DNA in the faecal or urine specimens with absence of any spores.
Besides, the false negative results recorded in the present work may reflect improper specimen handling, and may also indicate the presence of other DNA polymerase inhibitors like haem and bilirubin which can result in failure of proper DNA amplification (Da Silva et al., 1999; Franzen and Muller, 1999; Accoceberry et al., 2001). It has to be noted that no testing for PCR inhibitors was done in the present study, thus we cannot exclude the possibility of presence of inhibitors in the used samples. Furthermore, Wittwer et al. (1997), Nath et al. (2000) and Karsai et al. (2002) stated that in SYBR Green PCR, the dye used appears to be inhibitory to the PCR reaction which can be overcome by increasing the concentration of MgCl2. Also, the degradation products of the colouring agent used in PCR have also been reported to be inhibitory to the amplification reaction (Karsai et al., 2002).
Using melting curve analysis in SYBR Green real-time PCR, two cases were diagnosed as unspecified species as their amplification plot showed single band of amplification with single amplicon, and single melting temperature that was very close to the range of the melting temperature of the four targeted species. The recorded size of those two cases by the micrometre in the stained stool samples were in the size range of E. bieneusi and E. intestinalis so they could be mutations from those species. Since melting temperature depends on the GC/AT ratio, so any mutation could result in change in the melting temperature. This was agreed with Ririe et al. (1997). Moreover, the unspecified species could be one of the other Microsporidia species affecting humans as Nosema, Pleistophora, or Trachipleistophora species. However, this could be confirmed after determination of their sequences. This was in accordance with Polley et al. (2011), who reported similar results and suggested that the two unspecified species in their results could be diagnosed as Pleistophora species depending on their sequences which were close to the sequence of Pleistophora species.
When comparing the results of the parasitological and molecular studies regarding identification of Microsporidia species, we found that there was a statistically significant difference between the two methods in their capability of species identification. However, statistical analysis proved a fair agreement. Taking into consideration, the highly subjective measures taken by ordinary micrometre lens, in addition to the effort and experience needed to measure the spores, it appears mandatory to find an easier and more objective method. Thus, reference melting temperatures detected by using melting curves are easily read by fluorescent dye emission in SYBR Green real-time PCR which provide a brilliant easy method to categorize the type of microsporidial infection. Adding to this, there is no time consumption to identify the species of infection by our PCR method as it is a core step in diagnosis. Therefore in this study, we considered the SYBR Green real-time technique the gold standard for species identification and its results were analysed in relation to age, sex, and causes of immunosupression.
Our results showed that the majority of species detected by SYBR Green real-time PCR were E. cuniculi and E. hellem. However, in most of the previous studies, E. bieneusi was the major species detected, followed by E. intestinalis (Weber et al., 1994; Franzen and Muller, 1999; Graczyk et al., 2007). Nevertheless, our findings were in accordance with results of a study performed by Sak et al. (2011), in which E. bieneusi was detected less frequently than E. cuniculi by four times, while E. intestinalis was seldom identified.
It was documented that there is no significant effect of the age on the rate of Microsporidia infection. This is in accordance to Furuya (2009) and Turk et al. (2012). However, considering the type of microsporidiosis, previous studies showed a highly prevalent infection of E. bieneusi among young children aged from 0-36 months of age (Tumwine et al., 2002; Leelayoova et al., 2005). The results of these studies were coincided with ours.
Considering sex as a demographic factor influencing microsporidiosis, we found that females constituted higher percentage of infection with Encephalitozoon species than males. This could be attributed to the close contact of females with children who are at risk of continuous infection due to their improper hygienic and feeding habits. While in E. bieneusi, 70% of the infections were in males. Our reported results as regards E. bieneusi infection were agreed with Akinbo et al. (2013) who showed that the E. bieneusi infection rate was significantly higher in males (13.1%) than females (3.5%). Similar results were also obtained by Halanova et al. (2013) who observed that E. bieneusi infection is 4.3 times higher in males than females. The difference between both sexes for Microsporidia species predilection was statistically insignificant. This could be explained by the similar impact of low economic status, nutritional habits and sanitary conditions on both sexes.
Trying to find if the immune status of the host can influence the type of Microsporidia infection, we grouped the immunosuppressed patients included in the present study who were positive for microsporidiosis by real-time PCR according to their causes of immunosuppression. Inspite of the insignificant variation between causes of immunosuppression and species of Microsporidia, we found that; E. bieneusi was mainly abundant in patients with renal failure (40%), while leukaemia patients comprised 57% of those infected with E. hellem. Our results were agreed with Chabchoub et al. (2009) and Ghoshal et al. (2015) who reported that cancer patients had E. hellem as the commonest microsporidial infection, while E. bieneusi infection was more common to happen in HIV-infected patients.
To conclude our results, we observed that SYBR Green real-time PCR had the advantage of being a fast and reliable method. Within one hour of the PCR run, it allowed simultaneous detection and differentiation of microsporidial infections in large number of samples (up to 48 samples). On the contrary, staining of the same number of samples with MTS and considering the time needed for microscopic detection of positive samples and measuring spores could take few days. Furthermore, the SYBR Green method used did not require well trained experts to detect the type of infection.
Inspite of the higher expenses of the SYBR Green real-time PCR compared to the low cost of microscopic examination, the former could be considered as an accurate method for detection of infection. Commercial availability of the kits for routine use in clinical laboratories will be cost effective as it is nearly sure one step diagnosis not requiring further confirmation. Besides, it will save false positive cases detected by microscopy from the hazards of needless treatment. Moreover, it will guide clinicians for an appropriate treatment given for certain species to prevent the systemic complication of the drugs, especially in immunosuppressed patients.
Therefore, compared to light microscope, SYBR Green real-time PCR method was proved in our study to be a perfect step forward in diagnosis of intestinal microsporidiosis.
Acknowledgments
We gratefully acknowledge financial support from the Parasitology Department, Faculty of Medicine, Alexandria University.
conflict of interests
There exists no conflict of interest.
Authors’ Contribution
Prof. Dr. Eman El-Kerdany, Prof. Dr. Shwikar Ahmed, and Prof. Dr. Maha Gaafar contributed in inventing the idea of this article. All authors shared in conducting the practical part of this work and in writing this article.
References