Advances in Animal and Veterinary Sciences
Research Article
Characterization of Coa Gene and Antimicrobial Profiles of Staphylococcus Aureus Isolated from Bovine Clinical and Subclinical mastitis
Gamal Younis1, Asmaa Sadat*, Mona Maghawry2
1Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, Mansoura, 35516; 2Animal Health Research Institute, Dokki, Egypt.
Abstract | Staphylococcus aureus (S. aureus) is one of the major pathogens involved in bovine mastitis. Monitoring of antibiotic use could assess the hazard of S. aureus in cow’s milk. The objective of this study was to determine the prevalence of S. aureus in mastitic dairy cows in some localities in Dakahlia and Damietta Governorates, Egypt and to phenotypically characterize the antimicrobial susceptibility of the obtained isolates. In that context, a total of406 milk samples were collected from dairy cows suffering from clinical (n=100) and subclinical (n=306) mastitis from four different dairy farms. Milk samples were subjected to conventional bacteriological isolation techniques, and confirmed as S.aureus by using PCR assay targeting nuc (S. aureus-specific thermonuclease) and coa (coagulase) genes. Interestingly, 28 S. aureus strains tested negative to coagulase enzyme phenotypically were positive for Cao gene. The prevalence rate of S. aureus in clinical and subclinical mastitic milk samples was 66% (66/100) and 30.72% (94/306) respectively with an overall prevalence of 39.40% (160/406). All S.aureus isolates were tested against 10 different antimicrobials belonged to 7 antimicrobial classes. S.aureus strains exhibited a high rate of resistance to vancomycin (93.75%), penicillin (86.25%), trimethoprim (60%), and oxacillin (58.75%). A medium rate of resistance was observed to clindamycin (41.25%), ciprofloxacin (41.25%) and erythromycin (37.5%).On the other hand; S.aureus isolates displayed a low frequency of resistance to gentamicin (21.25%), ampicillin/sulbactam (20%) and chloramphenicol (18.75%). The multidrug-resistance to three or more classes of antimicrobials was detected in 108isolates (67.5 %). In conclusion, antimicrobial resistance of S.aureus was prevalent in dairy herds in the study area which represents a public health hazard. Hence, control and prevention policies should be implemented to minimize the dissemination of resistance trend.
Keywords | Antimicrobial resistance, Cows, Coa gene, Mastitis, Nuc gene, Staphylococcus aureus.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | March 03, 2018; Accepted | March 26, 2018; Published | April 18, 2018
*Correspondence | Asmaa Sadat, Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, 35516, Egypt; Email: [email protected]
Citation | Younis G, Sadat A, Maghawry M (2018). Characterization of coa gene and antimicrobial profiles of staphylococcus aureus isolated from bovine clinical and subclinical mastitis. Adv. Anim. Vet. Sci. 6(4): 161-168.
DOI | http://dx.doi.org/10.17582/journal.aavs/2018/6.4.161.168
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2018 Younis et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Bovine mastitis is considered as a serious threat in dairy industry as it causes huge economic losses in dairy herds (Awale et al., 2012; Xue et al., 2014). Bovine mastitis may caused by either contagious or environmental causes. Contagious mastitis is caused by bacterial or mycotic pathogens. However, environmental mastitis is the result of injury, bruising, chilling, or rough and improper milking. Large number of pathogens has been isolated from cases of mastitis in dairy cows. Mastitis causing pathogens have been classified as major and minor pathogens (Firaol et al., 2013).
Among the pathogen causing mastitis, S. aureus is one of the major pathogens of clinical and subclinical bovine mastitis (Fox et al., 2001; National Mastitis Council, 2004; Xue et al., 2014). Pathogenic S.aureus differs from other staphylococcus species by possessing coagulase enzyme and called coagulase positive staphylococcus (CPS) (Cunha, 2009). S.aureus in milk can affect the consumer in case of ineffective pasteurization or also in dairy products with raw milk; this represents an important public health hazard (Silva et al., 2013). It can also cause outbreaks of variety of diseases such as food poisoning (nausea, violent vomiting, and abdominal cramping with or without diarrhea) (Le Loir et al., 2003), osteomyelitis, septic arthritis or septicemia (Peles et al., 2007).
Antimicrobial therapy is an important tool in mastitis therapeutic programs. In the last decades, failure of antimicrobial therapy against Staphylococcus spp. has been recorded (Vintov et al., 2003). This therapeutic failure is associated with antimicrobial resistance of S. aureus, intracellular location of S. aureus or microabscess formation deep inside the udder tissues (Fox et al., 2001; National Mastitis Council, 2004). S. aureus strains have been observed for resistance against different antimicrobial agents including beta-lactams, aminoglycosides, fluoroquinolones, lincosamides, macrolides, andstreptogramins that are commonly used by veterinarians worldwide to treat mastitis (Hendriksen et al., 2008; Wang et al., 2008).
There is a great concern that the antibacterial-resistant agents in food animals can be transmitted to humans by dairy food chain (Irlinger, 2008).Therefore, this study was designed to determine the prevalence of S. aureus in mastitic dairy cows in Dakahlia and Damietta Governorates, Egypt and to genetically characterize the isolated strains using nuc (S. aureus-specific thermonuclease) and coa (coagulase) genes. In addition, the antimicrobial susceptibility patterns of isolated strains against common antimicrobial agents were also determined.
Materials and methods
Samples Collection
A total of 406 quarter milk samples were collected from 285 animals affected with mastitis. Samples were collected from four different dairy farms located at Dakahlia and Damietta Governorates, Egypt during the period between June 2016 and August 2017. Forms of mastitis were determined based on physical examination of the udder and California Mastitis Test (CMT) findings. Clinical mastitis was diagnosed based on the presence of abnormal changes in milk, mammary gland, teat, and systemic reaction (Quinn et al., 2004). Milk samples were collected aseptically. Quarters were washed with clean warm water and dried and the teat ends were disinfected with 75% ethyl alcohol then 8 to 10 ml milk samples were collected in sterile tubes after discarding the first three streams of milk. Milk samples were held in an ice box and transported immediately to laboratory for bacteriological analysis.
Bacteriological Examination
Milk samples were subjected to S.aureus isolation procedures based on the standard technique previously established by Wang et al. (2012). Briefly, milk samples were vortexed, and 10 µL of milk was plated on Baird-Parker agar plates (BPM, Oxoid, Basingstoke, UK) with 5% egg yolk and 1% potassium tellurite and incubated at 37°C for 24 h. For purification, one or two presumptive colonies per plate (black colonies surrounded by hallow zone) were transferred to trypticase soy agar (TSA; Oxoid, UK) plates. S.aureus isolates were subjected to Gram staining, coagulase test and the standard biochemical tests (Boerlin, et al 2003; De Freitas Guimarães et al., 2013). All isolates were stored in 20% glycerol solution at −80°C for further investigation.
Phenotypic Antimicrobial Susceptibility Testing
The susceptibility of S.aureus isolates to different antimicrobials was carried out using disc-diffusion method on Mueller-Hinton agar as recommended by CLSI (2013). Susceptibility of the isolates was determined against 10 antimicrobial agents (Oxoid, UK) including, ciprofloxacin (CIP, 5μg), erythromycin (E, 15 μg), gentamicin (CN, 10 μg), penicillin (P, 10 μg), clindamycin (DA, 2 μg), oxacillin (OX, 15 μg), Ampicillin-sublactam (SAM, 20 μg), chloramphenicol (C, 30 μg), trimethoprim-sulfamethazole (SXT, 25 μg), and vancomycin (VA, 30 μg). Interpretation of the results was done following CLSI guidelines (CLSI, 2012) (Table 3). S.aureus ATCC29213 (B-lactamase positive), and S.aureus ATCC25923 (B-lactamase negative) were used as quality control. Multidrug resistance was reported as a single isolate resistant to 3 or more antimicrobial classes (Waters et al., 2011).
Molecular Characterization of S.aureus
DNA extraction: Genomic DNA was extracted by boiling method (Alexopoulou et al., 2006). In brief, 3 to 5 bacterial colonies were picked up and suspended in 50 µl deionized water followed by boiling for 5 min and centrifuging at 10000 g for 1 min. The supernatant were then transferred and used as the DNA template for further molecular characterization.
PCR assay: PCR assay was performed for the detection of the nuc (encoding for the S. aureus-specific thermonuclease) gene and coa gene encoding for coagulase as previously described by Sallam et al. (2015) and Himabindu et al. (2009), respectively. The primers sequences (Hokkaido System Science Co.Ltd., Hokkaido, Sapporo, Japan and Metabion, Germany) and its amplicons size are listed in Table 1. Each PCR reaction was performed in a total vol
Table 1: Primer and cyclic PCR conditions for PCR amplification of various genes specific for molecular identification of Staphylococcus aureus
-
Target gene Primer direction and sequence Amplicon size (bp) Reference PCR conditions nuc F: GTGCTGGCATATGTATGGCAATTG
R:CTGAATCAGCGTTGTCTTCGCTCCAA
660 Initial denaturation:94˚C/2 minAmplification:(35cycles) Denaturation:98C / 10 s Annealing: 58C/30 s
Extension:68C/1min
Finalextension: 68C/7min
coa F: CGAGACCAAGATTCAACAAG
R: AAAGAAAACCACTCACATCA
500-1000 Initial denaturation: 94C/5 min Amplification:(35cycles) Denaturation:94C/30sec Annealing: 54C/1min
Extension:72C/1min
Finalextension:72C/10 min
ume of 25 μL, consisting of the following components: 12.5 μl master mix (Thermo scientific, USA); 1 μl of each primer; 2 μl DNA template and volume of the reaction mixture was completed to 25 μl using DNase/RNase-free water. PCR reactions were performed using 96 well Applied Biosystem,2720thermal cycler and cyclic PCR conditions described in Table 2 according to the referenced authors. Eight µl of each PCR product was separated by agarose gel electrophoresis, 1.2 % agarose prepared in TBE. Gels were stained with ethidium bromide and then visualized by Gel Doc (cleaver scientific ltd UV gel documentation system, USA).
Statistical Analysis
Differences between different antimicrobial agent in resistant, intermediate and susceptible Staphylococcus aureus strains were tested using the Chi square (χ2) test. χ2 was calculated 830.95 and the degree of freedom was 18. Differences among means with P < 0.000001 were considered as statistically highly significant.
Results
S.aureus Prevalence
The overall prevalence of S. aureus in this study was 39.4% (160/406) out of 406 milk samples from both clinical and subclinical mastitis. According to the type of mastitis, S. aureus was recovered at a prevalence rate of 66% (66/100) and 30.72% (94/306) from clinical and subclinical mastitis, respectively.
S. aureus isolates were identified with the nuc gene using PCR assay in all the biochemically identified isolates (Figure 1). Coagulase gene encoded by coa was also used for the confirmation of the isolated S. aureus, all the recovered S. aureus harbored the specific amplified products of coa gene at the expected size 500 bp except in ten isolates, in which two variants of coa gene were detected, one with the size of 500bp and the second variant was 600bp (two isolates), 700bp (four isolates), 800bp (two isolates) and 900bp (one isolate) (Figure 2). Amplification of coa gene showed that 28 isolates, which had been identified as coagulase negative staph aureus, contained coa gene (Table 2).
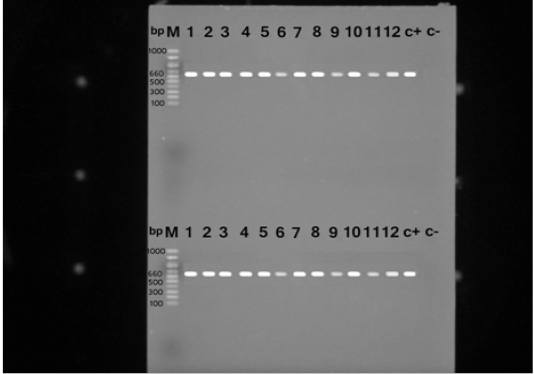
Figure 1: PCR products for nuc gene of Staphylococcus aureus strains showing amplified genes at the expected molecular size 660 bp from lane (1-12) and the last two for positive and negative controls, respectively.
Table 2: Difference between phenotypic and molecular typing of S. aureus
| No. of examined samples | No of S. aureus isolates | ||
| Coa +ve isolates | Coa – ve isolates | ||
| Coagulase test | 406 | 132 | 28 |
| Total: 160 | |||
| Coagulase gene amplification | 406 | 160 | -- |
| Total: 160 | |||
Antimicrobial Susceptibility Testing
Antimicrobial resistance was investigated against 10 antimicrobial agents. A high frequency of resistance was observed for vancomycin 93.75% (150/160), penicillin 86.25% (138/160), trimethoprim 60% (96/160), and oxacillin 58.75% (94/160). A medium frequency of resistance was observed for clindamycin 41.25% (66/160), ciprofloxacin 41.25% (66/160) and erythromycin 37.5% (60/160). A low frequency of resistance was displayed against gentamicin 21.25% (34/160), ampicillin/sulbactam 20% (32/160) and chloramphenicol 18.75% (30/160) (Table 3). The multidrug-resistance (resistance to three or more classes of antimicrobials) was detected in 108 isolates (67.5 %) (Table 4).
Table 3: Percentage of antimicrobial susceptibility
Antimicrobial agent Family Disc code CPD Resistance Intermediate Sensitive No % No % No % Penicillin β-lactamic
P 10 μg 138 86.25 22 13.75 Oxacillin OX 15 μg 94 58.75 22 13.75 44 27.5 Ampicillin-sublactame
SAM 20 μg 32 20 16 10 112 70 Ciprofloxacin Fluoroquinolone CIP 5 μg 66 41.25 60 37.5 34 21.25 Chloramphenicol Phenicols C 30 μg 30 18.75 66 41.25 64 40 Clindamycin Lincosamide DA 2 μg 66 41.25 94 58.75 Erythromycin Macrolide E 15 μg 60 37.5 96 60 4 2.5 Gentamicin Aminoglycoside CN
10 μg 34 21.25 22 13.75 104 65 Vancomycin Glycopeptide VA 30 μg 150 93.75 10 6.25 Trimethoprim-sulfamethazole Sulphonamide SXT 25 μg 96 60 22 13.75 42 26.25
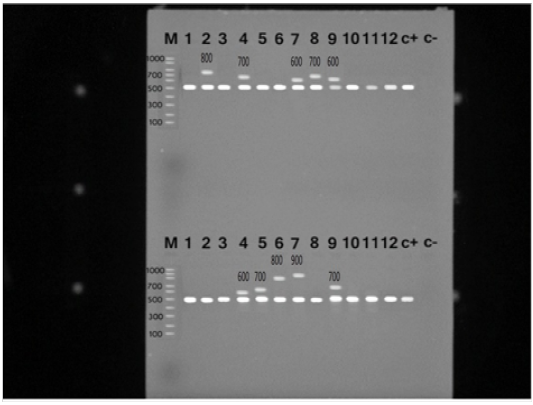
Figure 2: PCR products for coa gene of Staphylococcus aureus strains showing amplified genes at the expected molecular size 500-1000 bp from lane (1-12) and the last two for positive and negative controls, respectively.
Discussion
Staphylococcus aureus is a highly pathogenic micro-organism, which causes many serious diseases in both human and animals. It has been reported as the most commonly isolated micro-organism from cases of mastitis (Van Dui jkeren et al., 2004; Aires-de-Sousa et al., 2007). The present study was carried out to determine the prevalence of S. aureus in dairy cows affected with clinical and subclinical mastitis and to characterize the antibiotic resistance patterns. The recovered S. aureus isolates expressed the typical criteria of isolation which was consistent with those given by Boerlin et al. (2003).
In the present study, the overall prevalence of S. aureus was 39.4%, being higher in cows with clinical mastitis (66%) than those having subclinical mastitis (30.72%). Similarly, Asmelash et al. (2016) reported a high prevalence rate of S. aureus in clinical more than subclinical mastitis. However, other studies have proved that S.aureus is the main causative agent of subclinical mastitis (Gitau et al., 2014; Legesse et al., 2015). The overall prevalence of S. aureus in this study (39.4%) was in line with the prevalence (42%) recorded by Awad et al. (2017) in Egypt, Bedada and Adem (2011) and Lakew et al. (2009) in southern Ethiopia who recorded a prevalence of 39.1%, and 39.4% respectively. A relatively higher prevalence (54%) of S.aureus was recorded by Berchtold et al. (2014). Moreover, a higher prevalence (49.3%) was also recorded in Zimbabwe by Katsande et al. (2013),in South Ethiopia (51.2%) by Abebe et al. (2016), and in Brazil (53%) by Guimarães et al. (2016). However, lower prevalence than the present findings was reported by many authors all over the world (Li et al. 2009; Wang et al. 2014; Wang et al. 2016).
S. aureus isolates in this study were subjected to phenotypic and molecular characterization using the coa gene. Coagulase is a genetic marker for S. aureus strains responsible for coagulation of plasma and is considered as a significant virulence determinant of S. aureus (Watanabe et al., 2009). Among all S. aureus isolates in this study, 28 strains harbored coa gene, but tested negative with coagulase test. This suggests that coagulase gene is not functional in these 28 isolates. A similar finding of mutated coagulase has been reported previously (Phonimdaeng et al., 1990; Sunagar et
Table 4: Antimicrobial resistance patterns
-
Resistance pattern Total no of isolates % One antibiotic 20 12.5 VA 12 7.5 P 2 1.25 CIP 2 1.25 SAM 4 2.5 Two antibiotic 32 20 P,VA 26 16.25 OX, VA 2 1.25 DA,VA 2 1.25 P,DA 2 1.25 Three antibiotic 12 7.5 P,OX,VA 6 3.75 P,CIP,VA 2 1.25 P,VA,SXT 4 2.5 Four antibiotic 8 5 P,OX,VA,SXT 4 2.5 P,CIP,DA,VA 2 1.25 P,DA,VA,SXT 2 1.25 Five antibiotic 6 3.75
P,OX,CIP,VA,SXT 2 1.25 P,E,CN,VA,SXT 2 1.25 P,OX,CIP,VA,SXT 2 1.25 Six antibiotic 26 17.5 P,OX,DA,E,VA,SXT 4 2.5
P,CIP,DA.E,VA,SXT 2 1.25 P,OX,CIP,DA,VA,SXT 4 2.5 P,OX,SAM,CIP,CN,VA 2 1.25 P,OX,SAM,CIP,VA,SXT 6 3.75 P,OX,SAM,C,VA,SXT 8 5
Seven antibiotics 24 15 P,OX,CIP,DA,E,VA,SXT 16 10 P,OX,SAM,E,CN,VA,SXT 2 1.25 P,OX,C,DA,E,VA,SXT 2 1.25 P,OX,SAM,C,DA,VA,SXT 2 1.25
P,CIP,DA,E,CN,VA,SXT 2 1.25 Eight antibiotic 24 15 P,OX,CIP,DA,E,CN,VA,SXT 14 8.75 P,OX,SAM,CIP,E,CN,VA,SXT 2 1.25 P,OX,CIP,C,DA,E,VA,SXT 6 3.75
P,OX,SAM,DA,E,CN,VA,SXT 2 1.25 Nine antibiotic 4 2.5 P,OX,SAM,CIP,DA,E,CN,VA,SXT 2 1.25 P,OX,SAM,CIP,C,E,CN,VA,SXT 2 1.25 Ten antibioyic 2 1.25 P,OX,SAM,CIP,C,DA,E,CN,VA,SXT 2 1.25
al. 2013). Hence, proper identification of coagulase positive and coagulase negative species can’t be performed by phenotypic methods only but also, requires a combination of phenotypic and molecular assays (Akineden et al., 2011). Furthermore, confirmation of S. aureus was performed by PCR assay targeting nuc gene encodes the S. aureus-specific region of the thermonuclease which can degrade both DNA and RNA, and its enzymatic activity can resist 100°C for at least 1 h. PCR amplification for nuc gene is considered as an accurate, rapid, and safe screening method for S. aureus detection (Shrestha et al, 2002). Moreover, phenotypic methods are considered time consuming, expensive, and less accurate method so molecular screening of S.aureus is reported to be more efficient than other phenotypic methods (Aras et al., 2012).
In this study, S. aureus isolates exhibited a high resistance to vancomycin (93.75%). But vanomycin resistance was not recorded against S. aureus isolates from mastitic cow’s milk (Kumar et al., 2010; Wang et al., 2014). In addition, the high resistance of S. aureus to oxacillin (58.25%) contradicted the results obtained by many other authors (De Oliveira et al., 2000; Gentilini et al., 2000; Guler et al., 2005; Kumar et al., 2010) who reported susceptibility of S.aureus to oxacillin.
Concerning penicillin resistance, a higher rate of resistance was revealed in this study (86.25%). This finding went parallel with those reported in many studies (Guler et al., 2005; Turtoglu, et al., 2006; Li et al., 2009; Shi et al., 2010; Wang et al., 2014; Akindolire et al. 2015). On the other side, much lower resistance was previously recorded by others (Kumar et al., 2010; Haran et al., 2012). The variety in penicillin resistance between investigations could be attributed to many reasons including the differences in animal production systems and the use of antimicrobial drugs in each country (Guler et al., 2005).
Comparing to the results of the current study, Kumar et al. (2010) reported a low resistance to erythromycin (22.7%), and clindamycin (14.1 %) but relatively high resistance against gentamicin (30.5%), and almost the same rate of resistance to chloramphenicol (17.9). In other investigation conducted by Wang et al. (2014), a higher resistance to erythromycin (61%) and a lower resistance againsttrimethoprim/ sulphamethoxazole (58.6%), ciprofloxacin (15%), gentamicin (7.3%), and chloramphenicol (0.00%) was exhibited by S. aureus isolates. Moreover, a lower resistance was revealed for trimethoprim-sulfamethoxazole and erythromycin (Guler et al., 2005; Haran et al., 2012).For clindamycin and ciprofloxacin resistance, a high rate of resistance against these antimicrobial agents was also observed by Wang et al. (2008).
Taken together, it can be inferred that the isolated S. aureus had a high resistance to the antibiotics which are frequently used in the country. Besides, bovine multidrug resistant S. aureus strains could be zoonotic pathogens (Soares et al., 2012). Given that Vancomycin and oxacillin are important antibiotics for human, isolates.Our results indicated high resistance rate for oxacillin which is responsible for methicillin resistance. This may be an indication for contamination or even infection by human isolates (Spanu et al., 2012). Moreover, the difference of animal welfare system and the medication by the antimicrobial drugs caused this high prevalence of antimicrobial resistance (Guler et al., 2005). The antimicrobial agents being used for treatment of mastitic cases were unexpected to possess this increased resistance (Goni et al., 2004). This may be attributed to modification of the antibiotics by modifying enzymes (Goni et al., 2004) or the influence of excessive use (Turutoglu et al., 2006).
Conclusions
The widespread use of antibiotics on dairy farms and other food-producing animals could lead to emergence of antibiotic-resistant bacterial strains which represents a serious public health problem because of the possibility of dissemination of the antimicrobial-resistant bacteria to humans via food. Therefore; all possible control and prevention policies should be implemented to minimize the dissemination of resistance trend.
Acknowledgements
We gratefully acknowledge Dr. Eman Abo El-fadl at the Department of Animal Husbandry and Development of Animal Wealth, Faculty of Veterinary Medicine, Mansoura University for her technical help in this study.
Conflict of interest
There is no conflict of interest.
Authors Contribution
G. Y. designed the experiment and revised the manuscript; A. S. performed all the practical part, analyzed all the data, wrote the manuscript, revised the manuscript and was the corresponding author; M. M revised the manuscript. All authors approved the final version of the manuscript for publication.
References






