Advances in Animal and Veterinary Sciences
Research Article
Matrix Assisted Laser Desorption Ionization – Time of Flight Mass Spectrometry Proteomic Based Identification and Characterization of Pathogens Associated with Bovine Mastitis
Jayashri Shankar Rohokale, Prashant Patilba Mhase, Dushyant Madhukar Muglikar, Bhupesh Purushottam Kamdi*, Anilkumar Sadashiv Bannalikar, Ashok V Bhosale, Rahul P Kolhe
Department of Veterinary Microbiology, Krantisinh Nana Patil College of Veterinary Science, Shirval, Dist – Satara (MS), Maharashtra Animal and Fishery Sciences University, Nagpur, Maharashtra - 412801, India.
Abstract | In the present study bovine milk samples (n = 235) were screened for mastitis with California Mastitis Test (CMT) from Pune and Satara districts of western part of India. It showed prevalence of subclinical mastitis and clinical mastitis to be 60.40% and 2.20% in Pune district whereas, 94.4% and 9.9% in Satara district, respectively. CMT positive samples were inoculated on cultural media and 203 isolates either as single or combined associated with udder infections were generated. All these bacterial isolates were analyzed using MALDI-TOF-MS and the results were compared with classical biochemical methods and PCR assay, specifically for Staphylococcus aureus and E coli. Selective plating and biochemical identification yielded 80 (39.41%) Staphylococcus aureus isolates followed by E. coli 53 (26.11%) while remaining 70 (34.48%) were other minor 32 (15.76%) gram positive and 38 (18.72%) gram negative organisms. Furthermore, S. aureus as well as E. coli organisms were also confirmed by PCR assay by using specific primers targeting FemA gene and 16SrRNA gene, respectively. The results of MALDI-TOF MS technique proved equally (100%) effective in identification of both the species with highest accuracy as compared with bacterial culture and PCR assay, thus demonstrating the receipt of accurate reliability of MALDI-TOF MS in bacterial identification. Additionally, identification of minor mixed bacterial culture illustrated its increased additional utility. In brief, result demonstrated that MALDI-TOF MS can be used in veterinary diagnostic laboratories for identification of different bacterial species involved in mastitis. Hence, efficient animal management strategies for disease and milk quality control in the dairy industry can be implemented.
Keywords | Mastitis, Bacterial cultural, PCR, MALDI-TOF MS
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | June 08, 2016; Accepted | July 07, 2016; Published | July 14, 2016
*Correspondence | Kamdi Bhupesh Purushottam, Assistant Professor, Krantisinh Nana Patil College of Veterinary Science, Shirval, Dist – Satara (MS), India; Email: [email protected]
Citation | Rohokale JS, Mhase PP, Muglikar DM, Kamdi BP*, Bannalikar AS, Bhosale AV, Kolhe RP (2016). Matrix assisted laser desorption ionization – time of flight mass spectrometry proteomic based identification and characterization of pathogens associated with bovine mastitis. Adv. Anim. Vet. Sci. 4(7): 381-388.
DOI | Http://dx.doi.org/10.14737/journal.aavs/2016/4.7.381.388
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Rohokale et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Mastitis is a complex disease with multiple etiologies, causing inflammation of parenchyma of mammary glands, characterized via physical, chemical and frequently bacteriological alterations in milk as well as pathological changes in glandular tissues. It has evolved as a disease of highest economic significance particularly to the dairy industry worldwide and has great public health significance because of the ability of S. aureus to produce enzymes and enterotoxins which cause serious food poisoning (Benic et al., 2012). In dairy industry mastitis contributed to annual loss of approximately 2 billion dollars from USA and 526 million dollars from India (Hamadani et al., 2014). More than 200 microbial species, sub-species and serotypes have been isolated from bovine mammary glands and identified as causative agents associated with mastitis (Benhamed and Kihal, 2006). The most common contagious pathogen involved in mastitis is S. aureus and Streptococcus agalactiae (Behiry et al., 2014). Other pathogens involved in mastitis includes E. coli, Streptococcus uberis, Streptococcus dysagalactiae, Klebsiella spp., Corynebacterium spp., Serratia, Pseudomonas, Proteus, Mycoplasma spp. and various fungal agents (Radostitis et al., 2000). The most frequently used diagnostic methods for detection of mastitis are Somatic Cell Count, California Mastitis Test while the gold standard method is bacteriological culture. However, there are several disadvantages associated with bacterial culture such as high number of leukocytes or presence of preservatives or residual therapeutic antibiotics yielding no bacterial growth from samples of truly sub-clinically infected cows (Koskinen et al., 2010). Use of molecular methods in detection and characterization of pathogens are developed over a period of last few decades. Since then for detection of a number of mastitis pathogens, PCR based techniques with high sensitivity and specificity have been described by many workers (Mehrotra et al., 2000; Mohamed et al., 2013), using which, most of the bacterial species can be identified in hours, however, it needs days for classical culture methods. Both biochemical and genetic analyses are truthful, but their application may be time consuming and comparatively costly affair (Castelani et al., 2013). Therefore microbiologists have concentrated their attention on cheaper and also fast but accurate technique of Mass Spectrometry (MS) for microbial identification, specifically Matrix Assisted Laser Desorption Ionization Time of flight Mass Spectrometry (MALDI-TOF MS) (Hillenkamp et al., 1991). It requires smaller amounts of biological material and involves simpler sample preparation protocols without primary assessment, such as Gram staining. Many microorganisms have been identified by MALDI-TOF MS via characteristic “chemical signatures” in a high throughput mode using simple extraction and sample work-up protocols. MALDI-TOF MS can be used for bacterial determination in a few minutes using crude bacterial extracts or lysate supernatants of whole cells from initial bacterial growth which requires on an average one day (Alispahic et al., 2010; Barreiro et al., 2010; El-Bouri et al., 2012). MALDI deals with thermolabile, non-volatile organic compounds and those of high molecular mass. It is used for the analysis of proteins, peptides, glycoproteins, oligosaccharides and oligonucleotides of pathogens (Hillenkampet al., 1991). In MS method DNA is generally detected by its negative ions, i.e. in the singly or multiple deprotonated form. Recently, several research groups have reported on the analysis of bacterial DNA and on PCR with subsequent MS analysis for the characterization of bacteria (Santos et al., 2013). Major thrust area in dairy sector needed to be addressed on priority is mastitis control and effective treatment with the objective of earliest possible identification and characterization of major pathogens, their epidemiology including molecular epidemiology and development of sustainable future strategies against pathogens of udder.
Therefore, this study was designed to identify the various bacteria present in the milk samples of dairy animals with MALDI-TOF MS keeping results of cultural and PCR techniques as reference for detection of S. aureus and E. coli. Simultaneously, it was also attempted to detect other minor poly-microbial flora present in mastitic udder of dairy animals with MALDI-TOF MS.
MATERIALS AND METHODS
Samples
Milk samples (235) from nine different dairy farms located at different geographical locations in Satara and Pune district of India, were collected and tested during the present investigation. Before collection of samples; teats were cleaned with warm water and dried and few strips were discarded followed by collection of 10 to 15 mL of milk. The teats were then scrubbed with paper towel containing 70% ethanol and samples were collected in sterile screw cap plastic vials after simultaneous screening for mastitis by employing CMT as per Schalm et al. (1971). According to the changes of color and grade of gel formation, CMT results were interpreted as ‘negative’, ‘trace’, ‘+’, ‘++’, and ‘+++’, as described by Gitau et al. (2014). The milk samples showing ‘+’ and above results were immediately transported to the laboratory on ice. Milk samples were preserved in deep freezer until further processing.
Phenotypic Characterization
All the CMT positive milk samples were processed for isolation of bacteria associated with mastitis for their initial enrichment by inoculating loop-full of the sediment of centrifuged milk in Brain Heart Infusion (BHI) broth and incubating it at 37°C for 12 hours. For isolation of the major pathogenic organism S. aureus selective plating was performed on mannitol salt agar. The plates were incubated at 37°C for 24hr. Colonies showing characteristic yellowish appearance on agar were picked up and considered as presumptive S. aureus isolates. S. aureus were detected as per their morphological, cultural characteristics and biochemical tests (Quinn et al., 2004). Simultaneously all milk samples were enriched in Enterobacteriaceae enrichment broth and tubes were incubated at 37°C for 24 hr and selective plating was performed on Mac Conkeys medium and EMB medium. The plates were incubated at 37°C for 24 hrs. Characteristic colonies with metallic sheen on EMB agar showing lactose fermentations on MacConkey’s agar were picked up and considered as presumptive E. coli isolates, further confirmed by their morphological, cultural and biochemical characteristics (Quinn et al., 2004). The pure colonies of organisms identified as S. aureus and E. coli were sub cultured in duplicate on nutrient agar slants and stored in refrigerator at 4°C until further use. From
Table 1: Nucleotide sequence and anticipated size of PCR products for S. aureus and E. coli specific oligonucleotide primer
|
Gene |
Primer |
Oligonucleotide sequence |
Product size |
|
femA |
GFEMAR GFEMAR |
(F) 5’ AAAAAAGCACATAACAAGCG3’ (R) 5’ GATAAAGAAGAAACCAGCAG 3’ |
132 bp |
|
16 S rRNA |
ECO 235 ECO 455 |
(F)5’ CACTATCGGTCAGTCAGAG 3’ (R) 5’ ATCAACCGAGATTCCCCCAT 3’ |
232 bp |
the BHI broth 0.1 ul of the cultures were also inoculated on BHI agar plates by spread plate method with sterile L-shaped loops and incubated at 37°C for 24 hours. All the individual colonies of organisms generated except identified as S. aureus and E. coli were then observed microscopically and their conventional characterization was attempted up to genus and presumptive species level. Pure colonies were sub cultured on nutrient agar slants and stored at refrigeration temperature until further processed with MALDI.
Molecular Detection
Fresh isolates from bovine mastitis identified by cultural technique as S. aureus and E. coli were further confirmed with molecular technique by performing genus specific Polymerase Chain Reaction (PCR) on the purified DNA of organisms. Purification and extraction of bacterial genomic DNA was performed as per standard protocol described by Arora et al. (2006). Extracted DNA templates were subjected to genus specific PCR assay. Identification of S. aureus was performed by using specific primers targeting FemA gene in PCR assay as per Mehrotra et al. (2000). E. coli were subjected for identification by using specific primers targeting 16SrRNA gene per Sabat et al. (2009). The PCR reactions were performed in a Veriti® 96-well Thermal Cycler (Applied 122 Biosystems, Foster, EUA) in a final volume of 25 μl in 0.2ml PCR tubes. The reaction mixture consisted of 5 μl of the DNA template from the bacterial cultures, 12.5 μl of master mixture with and 1μl each forward and reverse primers and final volume of 25 μl was achieved by adding molecular grade nuclease free water. For FemA gene, PCR mixtures were denatured for 5 minutes at 94°C and subjected to 30 cycles of amplifications (5 minute denaturation at 94°C, 2 minute annealing at 57°C and 7 minute extension at 72°C). A final extension step was done at 72°C for 7 minutes. Similarly for 16SrRNA gene, PCR mixtures were denatured for 5 minutes at 95°C and subjected to 35 cycles of amplifications (1 minute denaturation at 95°C, 45 sec annealing at 64°C and 1 minute extension at 72°C). A final extension step was done at 72°C for 7 minutes. The presence of specific length amplicons were confirmed by 1.25% agarose gel electrophoresis. The PCR products were stored in deep freeze until further use (Table 1).
Detection of Organisms with MALDI-TOF MS
Various types of isolates of microorganism were identified using MALDI-TOF MS performed on Microflex LT device (Bruker Daltonics GmbH, Bremen, Germany) with Flex Control (version 3.0) software (Bruker Daltonics) for the mechanical achievement of mass spectra in the linear positive mode in a range of 2 to 20 kDa, according to the instructions of the manufacturer. To prepare the samples for MALDI-TOF MS, the bacterial cultures were thawed, subcultured on fresh media and incubated for 24 hr in a brain heart infusion broth. The broths were centrifuged and pellets were mixed with 75% ethanol and washed by centrifugation. Supernatant was discarded and pellet were again suspended in fresh 75% ethanol solution and centrifuged (all centrifugation steps were at 15,000 × g for 2 min), and the supernatant was discarded by carefully pouring it from the microtube. The remaining pellet was carefully removed with a micropipette tip. Bacterial pellets were allowed to air dry at room temperature for 5 to 10 min. The 70% formic acid solution was added proportionally to the size of pellet to completely dissolve it. Subsequently, 100% acetonitrile was added to each sample in volumes equal to the 70% formic acid solution added, thus producing a bacterial extract in a 1:1 ratio of 70% formic acid and acetonitrile. A final centrifugation step was performed to separate bacterial cell debris from the supernatant containing the inner-cell proteins used for the MALDI-TOF MS identification (Ryzhov and Fenselau, 2001).
Twenty five isolates each of presumptively identified as S. aureus and E.coli by conventional identification via culture, and molecular identification by PCR technique were processed by MALDI-TOF MS for their confirmation. Other partially identified by cultural method and phenotypically unidentified colonies of 70 organisms isolated from milk samples on BHI medium were subjected for identification only with MALDI-TOF MS as per Behiry et al. (2014). For automated data analysis, raw spectra were processed using the MALDI Biotyper OC 3.1.66 software (Brucker Daltonic GmbH, with flexcontrol software, Leipzig, Germany) at default settings. The software performs normalization, smoothing, baseline subtraction, and peak picking, creating a list of the most significant peaks of the spectrum (m/z values with a given intensity, with the threshold set to a minimum of 1% of the highest peak and a maximum of 100 peaks). To identify unknown bacteria, each peak list generated was matched directly against reference libraries (4,623 species) using the integrated pattern-matching algorithm of the Biotype OC 3.1.66 software. The unknown spectra were compared with a library of reference spectra based on a pattern recognition algorithm using peak position, peak intensity distributions, and peak frequencies. Once a spectrum has been generated and captured by the software, the whole identification process was performed automatically, without any user intervention. The peak intensity ≥1.5 was considered as the secure genus and probable species level for detection of bacteria.
Table 2: Comparative prevalence of mastitis by CMT and Cultural test
|
Distt |
Farms Tested |
Milk sample |
CMT Test |
Cultural Test |
Agreement |
||
|
(n) |
% |
(n) |
% |
% |
|||
|
Pune |
2 OF |
94 |
60 |
63.8 |
53 |
56.4 |
88.33 |
|
2 UF |
40 |
21 |
52.5 |
21 |
52.5 |
100 |
|
|
Total |
134 |
81 |
60.5 |
74 |
55.2 |
91.30 |
|
|
Satara |
1 OF |
14 |
3 |
21.4 |
03 |
21.4 |
100 |
|
4 UF |
87 |
52 |
59.8 |
50 |
57.5 |
96.15 |
|
|
Total |
101 |
55 |
54.5 |
53 |
52.5 |
96.36 |
|
|
Grand total |
235 |
136 |
57.9 |
127 |
54 |
93.38 |
|
OF: Organized farm; UF: Unorganized farm
RESULTS AND DISCUSSION
Results of the present study regarding prevalence of mastitis in dairy animals are depicted in Table 2, checked via CMT test. It revealed 136 (57.87%) sample positive for clinical and sub-clinical mastitis out of 235 milk samples from different dairy farms. Amongst CMT positive animals 13 (9.5%) presented the clear cut signs of clinical mastitis whereas sub clinical mastitis was revealed in 123 (90.5%) animals. Prevalence of mastitis in Pune district was 60.48% and in Satara district it was 54.46% in cattle. On cultural examination 127 (54.04%) milk samples out of 235 resulted positive. Overall agreement of CMT results when compared with cultural test was 93.38% thus, indicating the well-known fact that along with CMT it is necessary to confirm mastitis cases by detecting the presence of actual pathogens in milk. Findings of mastitis prevalence in cross breed cattle were in concurrent with Mekibib et al. (2010) and Gitau et al. (2014). On cultural examination of 127 CMT positive samples, total 203 isolates of specific major culturally typable and some culturally non typable bacterial species were generated on BHI medium with spread plate technique either as single or in mixed proportion of two to three organism types. Consequent upon selective plating and morphological and biochemical characterization 80 isolates were identified as S. aureus (39.41%) and 53 were identified as E. coli (26.11%) species. Other 70 (34.48%) minor pathogens attempted for their presumptive phenotypic identification morphologically and biochemically either up to genus or up to species level were classified roughly as minor gram positive 32 (15.76%) and gram negative 38 (18.72 %) groups and processed with MALDI. Overall findings revealed the higher incidence of S. aureus followed by E. coli as major pathogenic agents in majority of mastitis cases. Overall percent agreement of CMT test with cultural examination (93.38%) ranged between 88.33% to 100% in detection of bovine mastitis indicating very high sensitivity of CMT for detection of mastitis and our findings were in close agreement with Gianneechini et al. (2002) while they were in closely concurrence with Alekish et al. (2013) and Gitau et al. (2014) indicating the highest prevalence of S. aureus followed by E. coli and Streptococcal species in mastitis.
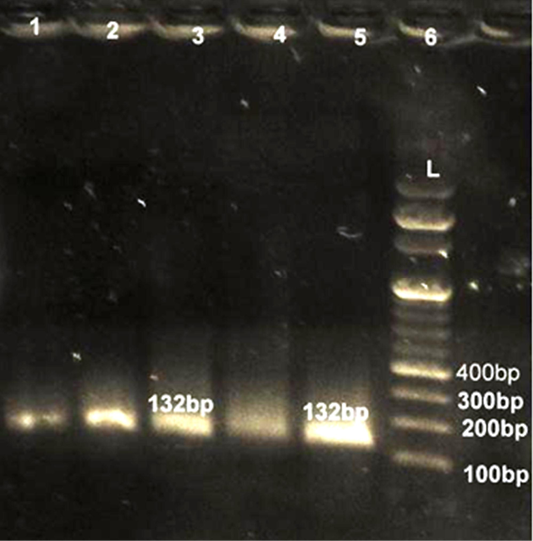
Lane 6: Marker Ladder of 100 bp size; Lane 1, 2, 3, 4, 5: Positive PCR product for femA gene (132 bp)
Molecular Detection of S. aureus and E. coli with Gene Specific PCR
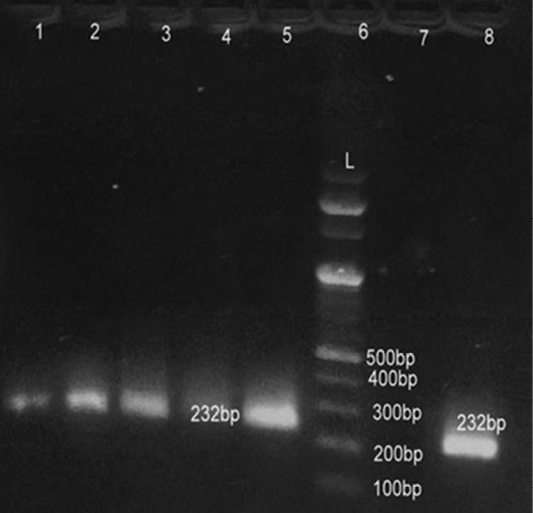
All 80 culturally identified colonies of S. aureus and 53 colonies of E. coli were processed for confirmation with gene specific PCR. For detection of S. aureus fragment of femA gene was amplified while for E. coli fragment of 16S rRNA was amplified in PCR assay. All 80 (100%) culturally identified S. aureus were specifically reconfirmed by generating 232 bp size band through agarose gel electrophoresis (Figure 1). Similarly all 53 (100%) purified DNA of E. coli species were also confirmed with 16SrRNA PCR yielding 132 bp size product on electrophoresis (Figure 2). Our results were in close agreement with Talib et al. (2009) employing pentaplex PCR for 16SrRNA and femA genes examining 230 clinical isolates and found to have 100% diagnostic accuracy. Previous findings of Sabat et al. (2009) were in agreement with present findings suggestive of 16SrRNA PCR as a robust and reliable method for identification of E. coli with highest sensitivity (100%).
Lane 6: Marker Ladder of 100 bp size; Lane 1, 2, 3, 5, 8: Positive PCR product for 16S r RNA gene (232 bp)
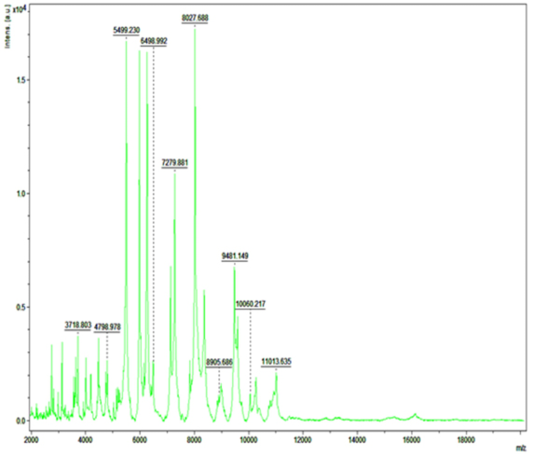
Detection of Bacteria Associated with Bovine Mastitis by MA LDI-TOF MS
In recent years, MALDI-TOF MS has been established for culture based pathogen identification in many clinical microbiology laboratories. Basically, in this technique dozens of microbial proteins as peaks with an exactly determinable mass to charge (m/z) ratio from whole cell mass spectra is analyzed. The observed degree of molecular mass conservation amongst these proteins renders spectral resemblance a suitable marker of phylogenetic kinship and enables existing commercially available fingerprinting systems to reliably infer species identity of unknown isolates from whole spectrum similarity comparisons with reference spectra (Christner et al., 2015). Direct MALDI-TOF MS analysis of bacterial cells requires can be accomplished within a few minutes as it require little effort for sample preparation. Therefore, MALDI-TOF MS is exposed to be a more significant and fast tool for the quick identification of diverse strains of S. aureus, Strep. agalactiae and CNS (Behiry et al., 2014). In earlier studies Wang et al. (2013) had identified 92 strains of S. aureus by MALDI-TOF MS from 100 milk samples at a secure genus and probable species level indicating the higher specificity of MALDI-TOF MS. In present research previous findings of cultural examination and gene specific PCR of S. aureus and E. coli were considered as reference and it was attempted to evaluate the accuracy of MALDI technique in identifying these two bacterial species.
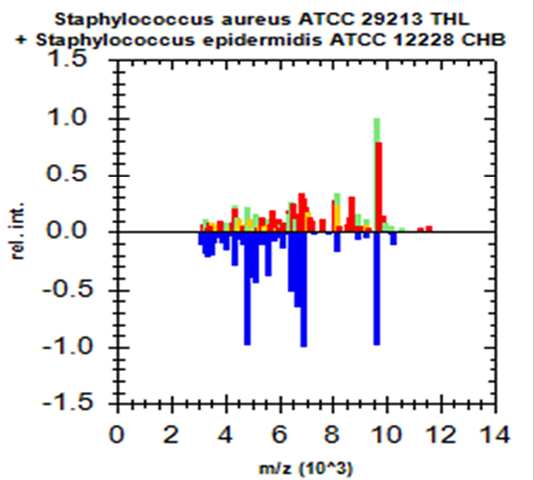
Green colour indicates good matching peaks; yellow indicates a moderate match; red indicates no match
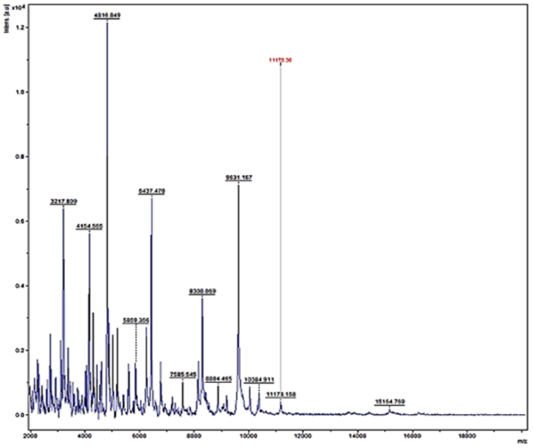
Randomly, 25 freshly grown colonies of each of these organisms were processed with MALDI-TOF MS and were identified with cent percent efficacy (100%) as S. aureus and E. coli within very short time. PCR analysis for other culturally isolated and partially identified organisms was not attempted during this research work because of their complexity in phenotypic identification and difficulty in availability of robust multiplex PCR technique for all of them. Hence, all such 70 isolates were straightway subjected for identification with MALDI and the results obtained were as depicted in Table 3. MALDI method could identify 55 (78.57%) of them with peak intensity of > 1.5 indicative of secure genus and probable species levelidentification (Figure 4 and 5).
Table 3: Detection of bacterial pathogens of bovine mastitis with MALDI –TOF MS (Score > 1.5)
|
Score Value |
Detected Species |
No. Tested |
No. Positive |
|
1.673-2.184 |
S. aureus |
25 |
25 |
|
1.606-2.010 |
E. coli |
25 |
25 |
|
1.964-2.010 |
Salmonella anatum |
70 |
4 |
|
1.699–2.010 |
Streptococcus dysgalactiae |
70 |
3 |
|
1.851-1.964 |
S. xylosus |
70 |
3 |
|
1.634-1.964 |
Enterobacter cloacae |
70 |
3 |
|
1.606–2.010 |
Proteus mirabilis |
70 |
3 |
|
1.559-1.606 |
Clostridium difficile |
70 |
3 |
|
1.730-1.789 |
Enterococcus faecium |
70 |
3 |
|
1.770-1.851 |
Stenotrophomonas sp. |
70 |
2 |
|
1.798-1.851 |
Klebsiella oxytoca |
70 |
2 |
|
1.870-2.010 |
S. hominis |
70 |
2 |
|
1.698-1.789 |
S. epidermidis |
70 |
2 |
|
1.717-1.851 |
S. cohnii |
70 |
2 |
|
1.634-1.964 |
S. schleiferi |
70 |
2 |
|
1.559-2.010 |
Pseudomonas fluorescens |
70 |
2 |
|
1.712-1.789 |
Proteus vulgaris |
70 |
2 |
|
1.584-1.789 |
Citrobacter freundii |
70 |
2 |
|
1.790-1.851 |
Streptococcus faecalis |
70 |
1 |
|
1.634-1.851 |
S. sciuri |
70 |
1 |
|
1.671-1.789 |
Raoultella ornithinolytica |
70 |
1 |
|
1.647-1.789 |
Pseudomonas azotoformans |
70 |
1 |
|
1.597-1.789 |
Cupria vidushector |
70 |
1 |
|
1.712-1.851 |
Clostridium beijerinckii |
70 |
1 |
|
1.690-1.789 |
Actinomyces urogenitalis |
70 |
1 |
|
1.634-1.851 |
Azoarcus communis |
70 |
1 |
|
1.559-1.634 |
Arthrobacter polychromones |
70 |
1 |
|
1.690-1.789 |
Acinetobacter pitti serovar 3 |
70 |
1 |
|
1.560-1.789 |
Lactobacillus agilis |
70 |
1 |
|
1.559-1.634 |
Lactobacillus aviaries |
70 |
1 |
|
1.597-1.789 |
Aeromonas encheleia |
70 |
1 |
|
1.712-1.851 |
Aeromonas molluscorum |
70 |
1 |
|
1.559-1.789 |
Camphylobacter jejuni |
70 |
1 |
|
< 1.5 |
Non typable |
70 |
15 |
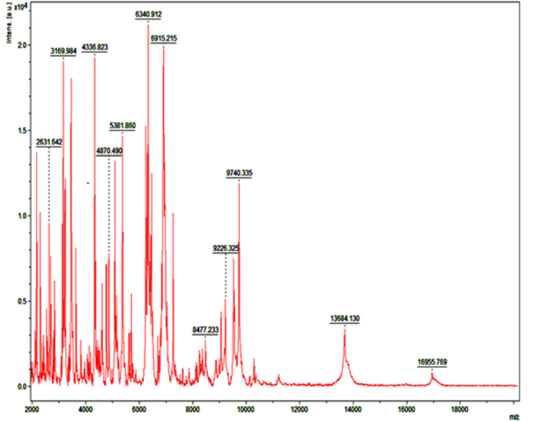
Figure 5: Mass spectra of E.coli by MALDI-TOF
Lower or no peak intensity values were observed for 15 (21.43%) isolates, therefore considered as non typable organisms. The Staphylococcal species culturally identified as minor micrococci were detected by MALDI as 3 isolates of S. xylosus (1.47%), 2 isolates each of S. cohnii , S. schleifer, S. hominis and S. epidermidis (0.99%, each) and 1 isolate of S. sciuri (0.49%). The Association of minor Staphylococcus spp. with milk of dairy animals was closely in accordance with Behiry et al. (2014). Other 8 gram positive Streptococcal species identified morphologically and culturally were confirmed as 4 isolates of Streptococcus dysgalactiae (1.97%), 3 isolates of Enterococcus faecium (1.48%) and 1 of Streptococcus faecalis (0.49%). Total 5 colonies of saprophytic gram positive rods in bovine milk identified upto genus level phenotypically and culturally were detected with MALDI as 3 isolates of Clostridium difficile (1.48%) and 1 isolate each of Clostridium bejierinckii and Actinomyces urogenitalis (0.49%, each).
The two colonies identified phenotypically up to genus level of gram positive Lactobacillus species were generated from mastitic milk samples which were confirmed as 1 isolate each of Lactobacillus aviaries and Lactobacillus agilis (0.49% each) with MALDI. The important minor gram negative organisms other than E. coli were phenotypically identified up to genus level and processed by MALDI were confirmed as 4 species of Salmonella anatum (1.97%), 3 species each of Enterobacter cloacae and Proteus mirabilis (4.2%, each), 2 colonies each of Pseudomonas fluorescans, Klebsiella oxytoca, Citrobacter freundii, Proteus vulgaris as well as saprophytic Stenotrophomonas spp., (0.99%, each) while 1 isolate each of very uncommon organisms found in milk, probably the environmental contaminants and opportunistic agents identified as Raoultella ornithinolytica, Pseudomonas azotoformans, Cupriavidus hector, Azoarcus communis, Arthrobacter polychromones, Acinetobacter pitti serovar 3, Aeromonas encheleia, Aeromonas molluscorum and Campylobacter jejuni (0.49%, each).
These results correlated with Barreiro et al. (2010) and Christner et al. (2015) indicating utility of MALDI-TOF MS as quick and successfully method for phenotypic profiling of microorganisms. Presence of such non-infectious polymicrobial agents in bovine milk of mastitic animals is probably indicative of poor hygienic practices and immunosuppression in these animals however, such correlation was drawn from circumstantial background and need to be addressed critically during future studies.
CONCLUSION
From these results it could be concluded that S. aureus was the major pathogen followed by E. coli in bovine mastitis. MALDI-TOF MS maybe adjudged as emerging tool for specific and early detection of poly microbial infections like mastitis in dairy animals. In addition to time consuming and costly methods of phenotypic identification and conventional gene specific PCR technique MALDI-TOF MS can be implemented as effective tool for rapid and accurate detection of major pathogens in bovine mastitis and establishment of the role of such uncommonly associated minor bacterial agents that needs to be addressed separately on large sample size in dairy animals reared in organized or as unorganized backyard livestock in Indian tropical conditions.
Acknowledgements
Authors are grateful to the Department of Microbiology, College of Veterinary Science, Udgir for generously providing the facility for MALDI-TOF MS typing of cultures.
CONFLICT OF INTERESTS
All Authors have declared no conflict of interest.
Authors’ Contribution
All authors contributed equally.
REFERENCES