Advances in Animal and Veterinary Sciences
Short Communication
Extracellular Enzymes and Toxins of Pseudomonas Aeruginosa Strains Isolated from Clinically Diseased Egyptian Cows
Gamal Younis1, Amal Awad1*, Mona Maghawry2, Fatma Selim1
1Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Mansoura University, Mansoura, 35516; 2Animal Health Research Institute, Dokki, Egypt.
Abstract | Pseudomonas aeruginosa produces large array of extracellular toxins that play a crucial role in virulence potential. In this study, the virulence profile of 35 clinical isolates of P. aeruginosa distinctly recovered from pneumonic cattle, mastitic milk samples, and wound infections was determined using phenotypic and genotypic methods. The overall incidence rates from pneumonic cattle, mastitic milk samples, and wound infections were 6%, 25% and 13.3%, respectively with a total incidence of 14%. Recovered isolates were then assayed for their enzymatic activity and the results revealed that out of these 35 isolates, 28 isolates produced haemolytic activity on blood agar plates, 27 isolates showed lecithinase enzyme activity on egg yolk agar media and 12 isolates showed proteolytic enzyme activity on brain heart infusion agar medium supplied with gelatin. Three virulent genes (las B, tox A and exoS) were screened by polymerase chain reaction (PCR) and the results showed that las B, tox A and exoS genes were amplified from 14.7%, 8.57%, and 17.14% of tested isolates, respectively. In conclusion, the present studies confirmed previous observations that the virulence of P. aeruginosa is multifactorial; however, the results indicate that certain exoproducts may play a more important role in certain types of infection.
Keywords | P. aeruginosa, Phenotypic, Genotypic, Virulent genes, PCR
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | June 11, 2015; Revised | July 29, 2015; Accepted | July 30, 2015; Published | August 07, 2015
*Correspondence | Amal Awad, Mansoura University, Mansoura, Egypt; Email: [email protected]
Citation | Younis G, Awad A, Maghawry M, Selim F (2015). Extracellular enzymes and toxins of pseudomonas aeruginosa strains isolated from clinically diseased Egyptian cows. Adv. Anim. Vet. Sci. 3(10): 522-526.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.10.522.526
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Younis et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Infections with P. aeruginosa have become an important clinical problem, which are difficult to eradicate by conventional antibiotic therapy. This has stimulated much interest in elucidating the mechanism of pathogenicity for this organism, including study of several extracellular secretory products as possible virulence factors (Albert et al., 2014). The pathogenesis of P. aeruginosa opportunistic infections is multifactorial, as suggested by the large number of cell-associated and extracellular virulence factors; some of these virulence factors help colonization, whereas others aid in bacterial invasion (Rocham et al., 2003; Deepak et al., 2013). Pseudomonas aeruginosa produces an extracellular toxins, these toxins include haemolysin, lecithinase, protease, elastase, collagenase enzymes, exotoxin A and exoenzyme S (Matthew, 1984; Khalifa et al., 2011).
P. aeruginosa haemolysin is the most potent toxigenic factor. The activity of this factor against macrophages is directly proportional to its haemolytic activity. The activity of other fractions against macrophages is more variable, but they may contribute in different ways to the development of infection once entry into the lung has been achieved. P. aeruginosa has two pathways to take iron, one of these pathway is hemolysin, it appears to be cytotoxic for most eukaryotic cells, so the hemolysin contribute to invasion through their cytotoxic effects on eukaryotic cells (Gadeberg et al ., 1983). The ability of a strain of P. aeruginosa to initiate respiratory infection may be related to the degree of haemolysin production (Al-Dujaili, 1976). Alveolar surfaces of lungs are covered with surfactants, made up mostly of lipids. phospholipase (lecithinase) has the potential to destroy lung surfactants and lung tissue, thus possibly causing bronchiectasis and atelectasis (Reynolds and Fick, 1979). P. aeruginosa secretes multiple proteases including elastase and alkaline protease that have been implicated as virulence factors. The proteases have tissue-damaging activity and are capable of degrading various plasma proteins, complement and coagulation factors (Galloway and Peters, 1990). Proteases probably play a part in localized pseudomonas infections such as keratitis, pneumonia and burn infection. When invasion and colonization have occurred, septicemia is established (Wretlind and Pavlovskis, 1981). P. aeruginosa produces two different ADP-ribosyltransferase toxins: ETA and exoenzyme S (Bodey et al., 1983; Bever and Iglewski, 1988). Exoenzyme S causes significant tissue damage in lung, burn and wound infections (Wolfgang et al., 2003). The highly toxic ETA is produced by the majority of P. aeruginosa strains and can inhibit eukaryotic protein biosynthesis at the level of polypeptide chain elongation factor 2, similarly to diphtheria toxin. ETA consists of two subunits; fragment A is catalytic, and fragment B is responsible for interaction with eukaryotic cell receptors. ETA is cytotoxic to numerous mammalian cells (Matthew, 1983).
Molecular techniques, such as polymerase chain reaction (PCR) are rapid and reliable, inexpensive and different than biochemical and serological tests for the identification of P. aeruginosa exotoxins (Katarzyne and Piotr, 2009; Amini et al., 2010). Nikbin et al. (2012) studied a rapid identification of P. aeruginosa exotoxins isolated from pulmonary tract, wound and burn samples based on PCR amplification of I lipoprotein (oprI) for detection of genus and L lipoprotein (oprL) for detection of species of this organism, also exotoxinA (toxA), exoenzyme S (exoS) and elastase (las B) were detected. The purpose of this study was directed to detect the phenotypic and genotypic virulence profiles of P. aeruginosa recent isolates from different clinical diseased cows.
A total of 250 samples were collected aseptically from various sources (100 mastitic cow’s milk, 100 nasal swabs from pneumonic cows, 20 lung tissues from pneumonic cows and 30 wound swabs from infected cows). All the samples were transported to the lab as quick as possible for bacteriological examination in asterile ice box. Samples were directly streaked on pseudomonas agar media supplied with (Cetramide 20% and Nalidixic acid 1.5%). The inoculated plates were incubated aerobically at 37°C for 24 hours. Suspected colonies were subjected to morphological and biochemical characters according to Cruickshank and McCarteney (1996). Toxins concentration and purification were done according to (Pinghui et al., 1973).
Haemolytic activity was measured on blood agar media, Circles (10 mm) were made on blood agar plates and punched out, then, supplied with a crude culture supernatant from P. aeruginosa and incubated at 37°C for 24 h. Ratio of diameter of colony within the circle to that of the translucent zone of hemolysis (in mm) was used as a hemolytic index to represent the extent of hemolytic activity by different P. aeruginosa isolates (Sachin et al., 2012). Haemolytic titer was detected according to Richard (1962).
Lecithinase production was detected by adding10% (vol/vol) egg yolk enrichment to Trypticase soy. A white precipitate around or beneath an inoculum spot indicated lecithinase formation (Alina et al., 2013). Detection of lecithinase activity was done according to (Habermann and Hardt, 1972).
Extracellular proteases was measured by placing a loopful of the culture growth of P. aeruginosa was placed in the center of the brain heart infusion agar media supplied with 1% gelatin. After inoculation, the plates were incubated at 37°C and observed daily for ten days. Extracellular protease detection was done after staining Coomassie blue (0.25%, w/v) in methanol-acetic acid-water 5:1:4 (v/v/v) for 1 h at 28°C (Vermelho et al., 1996).
DNA was extracted from P. aeruginosa isolates after grown in Trypticase-soy broth at 37°C for 24 hours and DNA was extracted by using a commercial available DNA isolation kit (Promega, U.S.), following manufacturer’s recommendations. Elastase enzyme (lasB), Exotoxin A (toxA) and exoenzyme S (exo S) genes were detected by PCR using primers selected on the basis of the published PAO1sequence (Stover et al., 2000). The PCR mixtures contained PCR buffer (10 mM Tris/HCL, 50 mM KCL, 1.5 mM mgcl, pH 8.3), 2.5μl of each dNTP, 1μl of each primer, 0.2 μl Tag DNA polymerase and 2μl DNA template. The DNA was amplified using the following protocol: 94°C for 3 min, 30 cycles of 94°C for 30 seconds, 57°C for 1 min and, 72°C for1 min and 30 seconds, and 72°C for 5 min. Each gene was amplified separately. PCR products were separated in 1% agarose gel for 1h at 100 V, stained with ethidium bromide and detected by UV transillunination (Katarzyne and Piotr, 2009). Amplified genes were identified on the basis of fragment size (Table 1).
Table 1: Sequences of oligonucleotide primers
|
Gene |
Primer sequence (5`-3`) |
Product (bp) |
|
las B |
las BF- GGAATGAACGAAGCGTTCTCCGAC |
284 |
|
lasBR- TTGGCGTCGACGAACACCTCG |
||
|
Tox A |
tox AF-CTGCGCGGGTCTATGTGCC |
270 |
|
toxAR-GATGCTGGACGGGTCGAG |
||
|
exo S |
exoSF-CGTCGTGTTCAAGCAGATGGTGCTG |
444 |
|
Exo SR- CCGAACCGCTTCACCAGGC |
P. aeruginosa produces a variety of extracellular products which possess potent biological activity in mammalian tissues (Liu, 1974). Hemolysin, Lecithinase enzyme, protease enzyme, elastase, exotoxin A and exoenzyme S have been studied to determine their role as virulence factors in Pseudomonas infection. A total of 250 samples were collected from different cilincally infected cows (100 nasal swabs, 100 mastitic milk samples, 30 wound swabs and 20 lung samples) and examined bacteriologically for presence of P. aeruginosa, a total of 35 P.aeruginosa isolates including (6 nasal swabs, 25 milk samples and 4 wound swabs) were recovered with an incidence of 6%, 25%, 13.3%, respectively with a total incidence 14% (Table 2). No P. aeruginosa isolates recovered from lung tissue samples. Hanaa (1997) isolated 50 positive strains of P. aeruginosa from different animal lesions of total 279 samples with an incidence17.0%. Results obtained in Table 3 pointed out that 28 (80%) of P. aeruginosa strains produced haemolysin enzyme and 7 (20%) were negative for its production on blood agar media. Moreover, high level of haemolysin production was recorded from P. aeruginosa strains isolated from nasal swabs (83.3%) and in milk samples (80%). Haemolytic activity was detected by estimation of the translucent zone of haemolysis surrounding the cultured colonies. Pinghui (1957) made a survey of hemolysin production among species of Pseudomonads, using the cellophane plate technique. SeveraL pseudomonads, both animal pathogens and non-pathogens, were found to produce hemolysin and it appears probable that hemolysin production cannot be used to differentiate species among pseudomonads. It was noted, however, that phenazine pigment producers, regardless of their pathogenicity to animals, tend to produce high titers of hemolysin.
Table 2: Pseudomonas aeruginosa strains recovered from different cow’s samples
|
Type of examined samples |
No. of examined samples |
Positive |
|
|
No. |
% |
||
|
Nasal swabs |
100 |
6 |
6 |
|
Milk samples |
100 |
25 |
25 |
|
Wound swabs |
30 |
4 |
13.3 |
|
Lung tissues |
20 |
0 |
0.00 |
|
Total |
250 |
35 |
14 |
Lecithinase enzyme and its activity was detected, they act synergistically to break down lipids and lecithin where in Table 3, 27 (77.1%) of P. aeruginosa strains recorded to be lecithinase positive. The obtained results demonstrated a variation in the level of lecithinase enzyme within P. aeruginosa strains; there were a high level in milk samples and nasal swabs. Absence of a positive reaction on egg yolk agar with P. aeruginosa may be due to lipase acting on the precipitate formed by its lecithinase activity. Choline and phosphorus were split from egg yolk lecithin in spite of the failure of this organism to produce a typical egg yolk reaction, while, the positive reaction was identified as the lecithinase activity resulted in the liberation of phosphorus and choline with precipitation of fat which was the source of the opalescence (Susumu and Pinghui, 1967).
All the isolated P. aeruginosa strains were examined for presence of protease enzyme as shown in Table 3, it is noticed that 12 (34.3%) of P. aeruginosa strains produced an extra-cellular proteases on brain heart agar with 1% gelatin through hydrolysis of gelatin. In this study, gelatin was used as a substrate as it is the most effective protease inducing substrate, have a high molecular weight protein, induces an increase in the protease production to degrade the substrate to an available form for the microorganism. Laila et al. (1986) and Vermelho et al. (1996) also detected the presence of extracellular proteases from microorganisms on agar plates. Michael et al. (1980) recorded that protease activity was present in 98% of the clinical isolates of P. aeruginosa.
PCR was used for detection of P. aeruginosa exotoxin A (toxA), exoenzyme S (exo S) and elastase enzyme. The obtained results demonstrated that 16 (45.7%) strains were positive for lasB, 3(8.57%) strains were positive for toxA and 6 (17.14%) strains were positive for exo S production (Table 3) and these results agree with Song et al. (2000) and Katarzyne and Piotr (2009). The obtained results demonstrated that production of high levels of haemolysin (80%), lecithinase (77.1%), protease (34.3%) and elastase enzyme (45.7%) of tested P. aeruginosa strains recovered especially from pneumonia and mastitis infections rather than wound infection. Also, virulence gens such as las B (100%)
Table 3: The number and percentage of strains positive for presence of screened virulence factors
|
Source |
Virulence factors |
|||||||||||
|
Haemolysin |
Lecithinase |
Protease |
las B |
tox A |
Exo S |
|||||||
|
No. |
% |
No. |
% |
No. |
% |
No. |
% |
No. |
% |
No. |
% |
|
|
Nasal swabs (6) |
5 |
83.3 |
6 |
100 |
6 |
100 |
6 |
100% |
2 |
33.33 |
1 |
16.6% |
|
Milk samples (25) |
20 |
80 |
18 |
72 |
4 |
16 |
8 |
32% |
- |
0.00 |
4 |
16% |
|
Wound swabs (4) |
3 |
75 |
3 |
75 |
2 |
50 |
2 |
50% |
1 |
25 |
1 |
25% |
|
Total (35) |
28 |
80 |
27 |
77.1 |
12 |
34.3 |
16 |
45.7% |
3 |
8.57 |
6 |
17.14 |
and tox A (33.33%) were detected mostly in strains isolated from nasal swabs, while, las B (32%) and exo S (16%)were detected mostly in strains isolated from milk samples (Figure 1, 2 and 3). In summary, the present studies confirmed previous observations that the virulence of P. aeruginosa is multifactorial; however, the results indicate that certain exoproducts may play a more important role in certain types of infection. Future studies will be conducted on these virulence genes to establish a relationship between the infections and P. aeruginosa isolated strains.
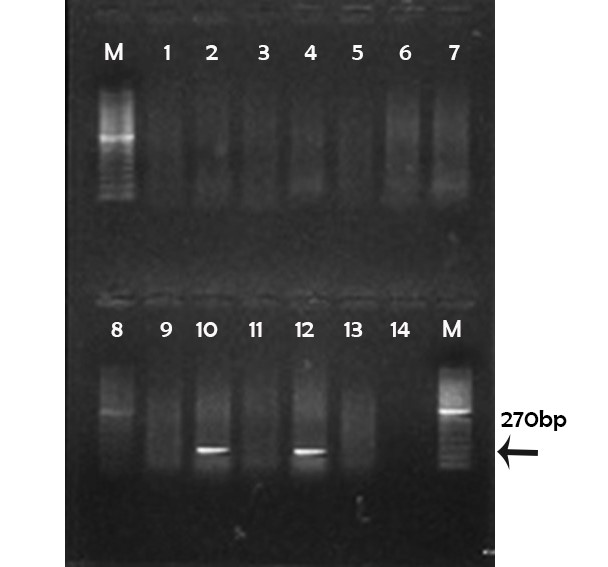
(M) marker 100pb; Lane 1, 2, 3, 4, 5, 6, 7, 8, 9, 11 and 13: negative; Lane 10, 12: positive; Lane 14: controle negative
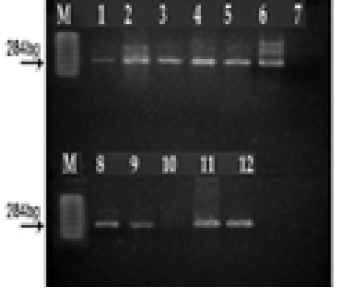
(M) marker 100 pb. Lane 1, 2, 3, 4, 5, 6, 7, 8, 9, 11 and12: positive; Lane 10: negative; Lane 7: controle negative
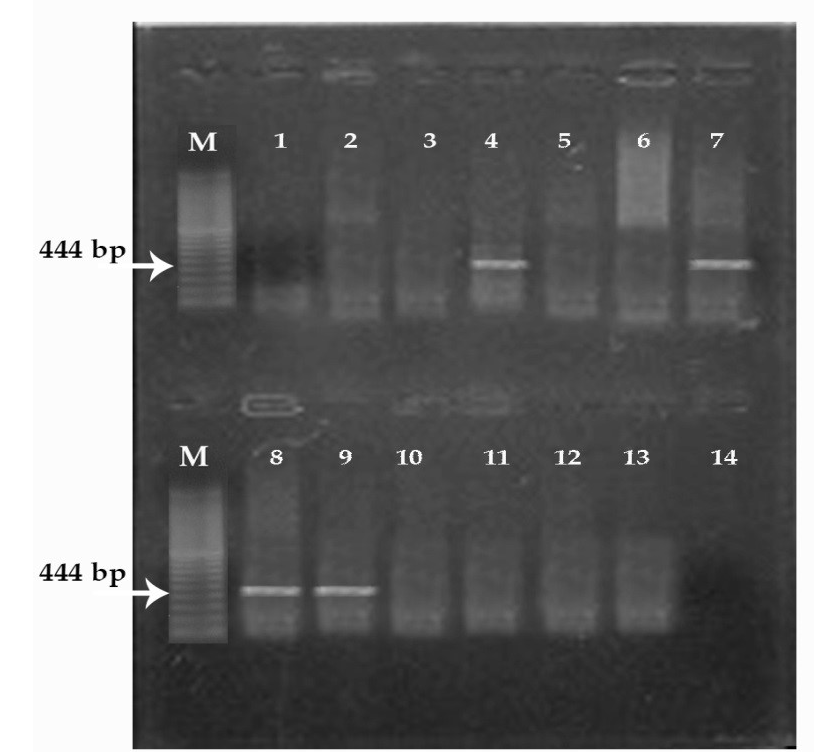
(M) marker 100 pb. Lane 1, 2, 3, 5, 6, 10, 11 and 12: negative; Lane 4, 7, 8, 9: positive; Lane 14: controle negative
Authors’ contributions
All the auhtors contributed equally in research activities and writing this article.
CONFLICT OF INTEREST
The authors state that there ia no conflict of interest.
References