Advances in Animal and Veterinary Sciences
Short Communication
Isolation of Antibiotic Resistant Salmonella Organisms from Aborted Cows and Emu Birds from District Ludhiana, Punjab, India
Vimlesh Gupta, Hari Mohan Saxena
Department of Veterinary Microbiology, College of Veterinary Science, GADVASU, Ludhiana, India.
Abstract | We investigated 102 samples (faecal swabs of live birds, spleen, caecum, liver and congested intestines of dead birds and uterine discharges, and diarrheal faeces of cattle, buffaloes and horses) for the presence of Salmonella organisms. Salmonella could be isolated from nine samples (7 from uterine fluid of aborted cows and 2 from fecal swabs of Emu birds). Identification of the isolates was done on the basis of cultural, staining and biochemical characteristics. All the 9 isolates were negative for oxidase and positive for catalase, motility indole lysine, and citrate utilization tests. The isolates were confirmed to be Salmonella by PCR using Salmonella genus specific oligonucleotide primers which yielded the desired amplicons of 496-bp. This indicates that Salmonella may also be associated with bacterial abortion and infertility in cattle. We found resistance to certain antibiotics among Salmonella and suggest here some of the most effective antibiotics for use in the field. The isolated organisms were found to be resistant to erythromycin and penicillin and sensitive to gatifloxacin, gentamicin, ciprofloxacin, norfloxacin, doxycycline, cefoparazone, ceftaxime, and oxytetracycline. Salmonellosis being a zoonosis, the findings are of public health significance as well.
Keywords | Salmonella, Abortion, Antibiotic Resistance, Cow, Emu
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | November 04, 2014; Revised | November 28, 2014; Accepted | November 29, 2014; Published | December 01, 2014
*Correspondence | Hari Mohan Saxena, College of Veterinary Science, GADVASU, Ludhiana, India; Email: [email protected]
Citation | Gupta V, Saxena HM (2014). Isolation of antibiotic resistant Salmonella organisms from aborted cows and Emu birds from District Ludhiana, Punjab, India. Adv. Anim. Vet. Sci. 2 (12): 652-656.
DOI< | http://dx.doi.org/10.14737/journal.aavs/2014/2.12.652.656
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2014 Gupta and Saxena. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Salmonella causes diarrhoea, dehydration, abortion, pneumonia, septic arthritis, meningitis, gangrene of distal extremities, and sudden death in domestic animals. It is a zoonotic disease which manifests as gastroenteritis, enteric fever, and a carrier state in most of the affected species. It decreases productivity and causes mortality in domestic animals, inflicting great economic losses to the dairy farmer, and is of public health significance.
The Centre for Disease Control and Prevention (CDC) reported that 9.4 million food borne illnesses, 55,961 hospitalizations and 1351 deaths occur in the United States each year (Scallan and Hoekstra, 2011). Salmonella is a major food borne bacteria which causes infection in humans as well as in food producing animals such as cattle, poultry and swine (Lynne et al., 2009; Guibourdenche et al., 2010). The infection of Salmonella is mostly due to contaminated food products from poultry, pigs and ruminants, contaminated drinking water or direct contact with infected animals (Mullner et al., 2009; Abdullah et al., 2010). In case of cattle and buffaloes it may cause abortion and infertility and leads to productive and economic loss (Verma et al., 1999). The organism may either cause abortion, or be associated with stillbirths or with the birth of normal healthy full-term calves (Hinton, 1971). Salmonella Enteritidis is an important cause of human illness with symptoms typically including fever, vomiting, diarrhea and abdominal cramps 12-72 hours after ingestion of the bacterium (CDC, 2010) and is responsible for millions of human cases of gastroenteritis and deaths worldwide (Majowicz et al., 2010).
Due to increase in use of antimicrobial agents in food animals as growth promoters, prophylactic agents and therapeutic remedies, increasing number of bacterial strains are gaining resistance (Zewdu and Cornelius 2009; Alexander et al., 2008). Nonspecific and non-judicious use of antibiotics for the bacterial agents aggravates such conditions. The number of antibiotic-resistant isolates identified in humans is steadily increasing, suggesting that the spread of antibiotic resistant strains is a major threat to public health (Dimarzio et al., 2013). The choice of right antibiotics is crucial to the control of the disease. We, therefore, analysed clinical samples from common domestic animals and poultry for presence of Salmonella and assessed the susceptibility of the field isolates of Salmonella to commonly used antibiotics to get the current status of antibiotic resistance in organisms in field conditions.
A total of 102 samples (faecal swabs of live birds, spleen, caecum, liver and congested intestines of dead birds and uterine discharges, and diarrheal faeces of cattle, buffaloes and horses) were collected. Samples were transported to laboratory on ice and processed within 1 hour. For isolation of organisms from clinical samples, first pre-enrichment and then enrichment were done in buffered peptone water and selenite cystein broth at 37°C for 18 hours and 24 hours, respectively. Selective plating on Hektoen Enteric Agar (HEA)/Brilliant Green Agar (BGA) plates was done by overnight incubation at 37°C as per Kaushik et al. (2014).
Identification of Salmonella isolates was done on the basis of cultural, staining and biochemical characteristics [H2S production, oxidase, catalase, motility indole lysine (MIL), and citrate utilization tests]. The genomic DNA of field isolates of Salmonella and the standard culture strain of Salmonella Enteritidis were extracted by the standard bacterial DNA extraction method as described by Wilson (1987). The DNA samples with OD260/280 value of ~1.8-1.9 were considered as pure and used for further analysis. The extracted DNA from Salmonella isolates and Salmonella Enteritidis were subjected to PCR by using Salmonella genus specific oligonucleotide primers of 25 base pairs as per Cohen et al. (1995). The primer sequences for the upper and lower oligonucleotides from 5’ to 3’, were as follows:
Lower strand- ACT GGC GTT ATC CCT TTC TCT GGT G;
Upper strand-ATG TTG TCC TGC CCC TGG TAA GAG A.
Antimicrobial agents (Himedia) viz. doxycycline, oxytetracyclin, enrofloxacin, penicillin, neomycin, amoxicillin, ampicillin, streptomycin, gentamicin and erythromycin were used for testing the sensitivity of the isolates. The culture plates were incubated micro aerobically at 37°C for 16-24 hours and zone of inhibition was measured for comparing the sensitivity and resistance of each organism.
A total of 102 samples comprising of fecal swabs of live birds, spleen, caecum, liver and congested intestine of dead birds and uterine discharges and diarrheal faeces of cattle, buffaloes and horses suspected of Salmonellosis were analysed (Table 1).
Salmonella could be isolated from 9 (7 from uterine fluid of cows and 2 from fecal swabs of Emu birds) out of 102 samples (Table 1). Light pink colonies of organisms on BGA and green colonies with black centre on HEA were observed. The organisms appeared as gram negative rods.
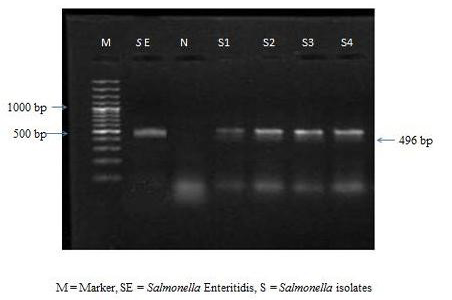
The colonies were subjected to biochemical tests like H2S production, catalase, oxidase, MIL and citrate utilization tests. All the 9 isolates were found to be negative for oxidase and positive for catalase, MIL and citrate utilization tests. All the culturally positive samples were further confirmed at molecular level by PCR. On amplification the extracted bacterial DNA yielded the desired amplicons of 496 bp (Figure 1).
Table 1: Isolation of Salmonellas from clinical samples of common domestic animals/birds
|
Animal/Bird |
Clinical symptom |
Type of sample |
No. of samples examined |
No. of negative samples |
No. of positive samples |
|
Poultry birds |
Diarrhea Congested intestine |
Faecal swabs Liver, caecum, spleen |
44 19 |
44 19 |
0 0 |
|
Emu birds |
Congested intestine |
Liver, caecum, spleen |
2 |
0 |
2 |
|
Cows |
Diarrhoea Abortion Dystocia |
Faecal sample Fetal stomach content, Uterine discharge Uterine discharge |
7 18 3 |
7 11 3 |
0 7 0 |
|
Buffaloes |
Diarrhoea Abortion |
Faecal sample Fetal stomach content, Uterine discharge |
2 2 |
2 2 |
0 0 |
|
Horses |
Diarrhoea |
Faecal sample |
5 |
5 |
0 |
Salmonella isolates were found to be 100% resistant to erythromycin and penicillin and sensitive to gatifloxacin, gentamicin, ciprofloxacin, norfloxacin, doxycycline, cefoparazone, ceftaxime, oxytetracycline. Sensitivity was 100% for gatifloxacin, gentamicin, ciprofloxacin, and norfloxacin, 77.78% for doxycycline, cefoparazone, and ceftaxime and 44.44% for oxytetracycline, respectively (Table 2).
Although Salmonella is associated with enteric infection and diarrhoea, particularly in calves, it may also cause infection in pregnant cows and may result in abortion which is the only clinical manifestation of infected herd. Colonization in reservoir hosts often occurs in the absence of clinical symptoms; however, S. enterica may cause acute enteritis or translocate from the intestines to other organs causing fever, septicaemia and abortion (Stevens et al., 2009). Rings (1985) opined that abortion due to Salmonella may be due to development of fever. In our study, 7 out of 18 samples from aborted cows and buffaloes, were positive for Salmonella. Thus, Salmonella may be a major cause of abortion in cows which leads to a great economic loss to the farmers.
Sanjrani et al. (2013) found that in case of mixed infection, abortions due to Brucella along with Salmonella were in highest frequency rather than other mixed in fections. Verma et al. (1999) investigated the role of salmonellosis in causation of abortions and infertility from 43 buffaloes and 110 cows. They isolated Salmonella Typhimurium from single abortion in a buffalo and 2 isolates of Salmonella Dublin from 7 aborted cows and 2 isolates of Salmonella Typhimurium from uterine discharges of 33 endometritic cows. They also observed that cyclic repeat breeding in buffaloes and cyclic repeat breeding, cervicitis and vaginitis in cows were also related to Salmonella infection. There may be incidence of dystocia, still births and retention of the placenta in case of Salmonella Dublin infection (Richardson, 1973). These finding are in concurrence with the findings of our present study.
Table 2: Susceptibility of Salmonella isolates to various antimicrobial agents
|
Antimicrobial agent |
Number of isolates analyzed |
Number (& percentage) of samples |
|
|
Positive |
Negative |
||
|
Gatifloxacin |
9 |
9 (100)† |
0 |
|
Gentamycin |
9 |
9 (100) |
0 |
|
Ciprofloxacin |
9 |
9 (100) |
0 |
|
Norfloxacin |
9 |
9 (100) |
0 |
|
Doxycycline |
9 |
7 (77.78) |
2 (22.22) |
|
Cefoparazone |
9 |
7 (77.78) |
2 (22.22) |
|
Ceftaxime |
9 |
7 (77.78) |
2 (22.22) |
|
Oxytetracycline |
9 |
4 (44.44) |
5 (55.56) |
|
Erythromycin |
9 |
0 (0) |
9 (100) |
|
Penicillin |
9 |
0 (0) |
9 (100) |
†Values in the parenthesis indicate percent positivity or negativity.
Our results have reconfirmed the earlier observa tions of widespread prevalence of antibiotic resistance among Salmonella and prompt for undertaking such surveillance from time to time in different regions to get a realistic picture of the problem of emergence of resistant pathogens and to enable selection of the most effective antibiotics for use in the field. Murugkar et al. (2005) have also reported varying degrees of resistance of their Salmonella isolates against doxycycline, ampicillin, amoxicillin, tetracycline, chlortetracycline, nitrofurantoin, chlortetracycline, kanamycin, cephalexin, nalidixic acid, chloramphenicol, trimethoprim, ciprofloxacin, gentamicin, enrofloxacin and norfloxacin. Irimie et al. (2010) isolated Salmonella cholerae suis spp. from liver and intestine which were resistant to all the antibiotics used. Resistance to multiple antibiotics could possibly be due to the misuse of antibiotics in the feed.
Based on the findings of the present study, we opine that Salmonella enterica is not only a typical enteric pathogen but it may also be a major cause of bacterial abortion in cattle.
References