Advances in Animal and Veterinary Sciences
Research Article
Antimicrobial Resistance and Virulence Genotyping of Different Salmonella Serovars Isolated from Chickens in Egypt
Aly M. Ghetas1*, Abdelbaki, M.M1, Hanaa S. Fedawy1, Dalia M. Sedeek1, M. A. Bosila1, Samy. A.A2, Nagwa S. Rabie1
1Poultry Diseases Department, National Research Centre, P.O. 12622, Giza, Egypt; 2Microbiology and Immunology Department, National Research Center, , P.O. 12622, Giza, Egypt.
Abstract | Twenty eight Salmonella strains representing 9 Salmonella serovars (S.Agama, S.Blegdam, S. Enteritidis, S.Gueuletapee, S.Infantis, S.Kentucky, S.Montevideo, S.Typhimurium and S.Virchow) were previously isolated, purified, and identified in our laboratory from freshly dead and diseased chickens suspected to infect with Salmonellosis. In the present study, antimicrobial resistance to 15 different antimicrobials and virulence genotyping to those Salmonella strains were performed. The significantly higher rate of resistance was detected against amoxicillin-clavulanic acid (AMC) and ampicillin (AMP) (85.7% and 78.5% respectively) comparing to the significantly lower rate of resistance detected against etapenem (ETP), gentamicin (GEN), ciprofloxacin (CIP), and norfloxacin (NX) (0 %, 0%, 3.5%, and 3.5% respectively). High resistance to cephalosporin antibiotics were also reported in this study. Resistance to 3 antimicrobials or more were identified in 17 out of 28 tested Salmonella strains. Interestingly, two S. Typhimurium strains were resist to 9 and 12 out of 15 antimicrobials used in this study. A multiplex Polymerase chain reaction (PCR) targeting 17 virulence genes was performed for virulence genotyping among different strains. Interestingly, all 17 virulence genes were detected in S. Infants and one strain of S. Agama. From a public health aspect, continuous resistance of Salmonella spp. to antimicrobials represent a serious public health hazard. Furthermore, Identification of virulence genes can help us to further understanding of Salmonella pathogenesis.
Keywords | Salmonella, Serovars, Antibiotics, Resistance, Multiplex PCR, Broilers.
Received | July 22, 2021; Accepted | August 02, 2021; Published | November 01, 2021
*Correspondence | Aly M Ghetas, Poultry Diseases Department, National Research Centre, P.O. 12622, Giza, Egypt; Email: [email protected]
Citation | Ghetas AM, Abdelbaki MM, Fedawy HS, Sedeek DM, Bosila MA, Samy AA, Rabie NS (2021). Antimicrobial resistance and virulence genotyping of different salmonella serovars isolated from chickens in Egypt. Adv. Anim. Vet. Sci. 9(12): 2124-2131.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.12.2124.2131
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Ghetas et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Salmonellosis is a major zoonotic foodborne disease causing mortalities, gastroenteritis, and/or septicemia in humans (Majowicz et al., 2010; Newell et al., 2010; Eng et al., 2015). Salmonella Enterica serovars are responsible of millions of enteric infections and thousands of human deaths annually (Balasubramanian et al., 2019). Contaminated poultry and eggs act as a main reservoir for Salmonellae (Antunes et al., 2016). Contamination of poultry meats and meat products are happened due to improper hygienic measure throughout plants during evisceration, cooling, packaging, and transport stages (Zhang et al., 2013). Salmonellae infect poultry causing clinical signs and high mortality especially in young chicks. Furthermore, it can be transmitted vertically from broiler breeder chickens to their progeny (Barbour et al., 1999; Lister, 1988). Control and prevention of Salmonellosis in poultry farm are depending mainly on using antimicrobials at therapeutic or prophylactic levels (Paudyal and Yue, 2019; Yue, 2016)
Table 1: Antimicrobials used in this study
| Antimicrobial agents | Conc. (mcg) | Diameter of inhibition zone (mm) | ||
| S* | I* | R* | ||
|
Amoxicillin/Clavulanic acid (AMC) |
30 (20/10) | ≥ 18 | 14-17 | ≤13 |
|
Ampicillin (AMP) |
10 | ≥ 17 | 14-16 | ≤13 |
|
Cefaclor (CF) |
30 | ≥ 18 | 15-17 | ≤14 |
|
Cefeprime (CPM) |
30 | ≥ 25 | 19-24 | ≤18 |
|
Cefotaxime (CTX) |
30 |
≥ 26 | 23-25 | ≤22 |
|
Chloramphenicol (C) |
30 | ≥ 18 | 13-17 | ≤12 |
|
Ciprofloxacin (CIP) |
5 | ≥ 21 | 16-20 | ≤15 |
|
Trimethoprim/sulphamethoxazole (COT) |
25 (1.25/23.75) | ≥ 16 | 11-15 | ≤10 |
|
Doxycycline Hydrochloride (DO) |
30 | ≥ 14 | 11-13 | ≤10 |
|
Ertapenem (ETP) |
10 |
≥ 22 | 19-21 | ≤18 |
|
Tetracycline (TE) |
30 | ≥ 15 | 12-14 | ≤11 |
|
Norfloxacin (NX) |
10 | ≥ 17 | 13-16 | ≤12 |
|
Gentamicin (GEN) |
10 | ≥ 15 | 13-14 | ≤12 |
|
Kanamycin (K) |
30 | ≥ 18 | 14-17 | ≤13 |
|
Streptomycin (S) |
10 |
≥ 15 | 12-14 |
≤11 |
*These letters represent susceptibility to antimicrobials: S= sensitive, I = intermediate, R= resistant
Table 2: Oligonucleotide primers sequences of target Salmonella spp. genes with amplicon sizes.
| Gene | Primer sequence | Amplicon size (bp) |
| spvB | F: CTATCAGCCCCGCACGGAGAGCAGTTTTTA | 717 |
| R: GGAGGAGGCGGTGGCGGTGGCATCATA | ||
| spiA | F: CCAGGGGTCGTTAGTGTATTGCGTGAGATG | 550 |
| R: CGCGTAACAAAGAACCCGTAGTGATGGATT | ||
| pagC | F: CGCCTTTTCCGTGGGGTATGC | 454 |
| R: GAAGCCGTTTATTTTTGTAGAGGAGATGTT | ||
| cdtB | F: ACAACTGTCGCATCTCGCCCCGTCATT | 268 |
| R: CAATTTGCGTGGGTTCTGTAGGTGCGAGT | ||
| msgA | F: GCCAGGCGCACGCGAAATCATCC | 189 |
| R: GCGACCAGCCACATATCAGCCTCTTCAAAC | ||
| invA | F: CTGGCGGTGGGTTTTGTTGTCTTCTCTATT | 1070 |
| R: AGTTTCTCCCCCTCTTCATGCGTTACCC | ||
| sipB | F: GGACGCCGCCCGGGAAAAACTCTC | 875 |
| R: ACACTCCCGTCGCCGCCTTCACAA | ||
| prgH | F: GCCCGAGCAGCCTGAGAAGTTAGAAA | 756 |
| R: TGAAATGAGCGCCCCTTGAGCCAGTC | ||
| spaN | F: AAAAGCCGTGGAATCCGTTAGTGAAGT | 504 |
| R: CAGCGCTGGGGATTACCGTTTTG | ||
| orgA | F: TTTTTGGCAATGCATCAGGGAACA | 255 |
| R: GGCGAAAGCGGGGACGGTATT | ||
| tolC | F: TACCCAGGCGCAAAAAGAGGCTATC | 161 |
| R: CCGCGTTATCCAGGTTGTTGC | ||
| iroN | F: ACTGGCACGGCTCGCTGTCGCTCTAT | 1205 |
| R: CGCTTTACCGCCGTTCTGCCACTGC | ||
| sitC | F: CAGTATATGCTCAACGCGATGTGGGTCTCC | 768 |
| R: CGGGGCGAAAATAAAGGCTGTGATGAAC | ||
| lpfC | F: GCCCCGCCTGAAGCCTGTGTTGC | 641 |
| R: AGGTCGCCGCTGTTTGAGGTTGGATA | ||
| sifA | F: TTTGCCGAACGCGCCCCCACACG | 449 |
| R: GTTGCCTTTTCTTGCGCTTTCCACCCATCT | ||
| sopB | F: CGGACCGGCCAGCAACAAAACAAGAAGAAG | 220 |
| R: TAGTGATGCCCGTTATGCGTGAGTGTATT | ||
| pefA | F: GCGCCGCTCAGCCGAACCAG | 157 |
| R: GCAGCAGAAGCCCAGGAAACAGTG |
F= forward, R= reverse
However, multiple antimicrobial resistant Salmonella strains developed due to haphazard use of antimicrobials at recommended doses or at sub-therapeutic doses which representing a public health hazard (Antunes et al., 2016; EFSA, 2013) . Thus, continuous monitoring of antimicrobial resistance have a high priority.
The severity of Salmonella infection in human and animals is depending on the presence of virulence genes (Ammar et al., 2016). Several virulence genes have been reported. Virulence genes encoded proteins such as: invA, orgA, prgH, spaN, tolC, sipB, pagC, msgA, spiA, sopB, lpfC, pefA, and spvB which responsible of adherence, invasiveness, entry to non-phagocytic cells, survival within macrophage, and growing within the host. Other virulence genes (sitC and iroN) were involved in iron acquisition, while cdtB was responsible of toxin biosynthesis (Skyberg et al., 2006). Thus, detection of virulence genes among different Salmonella serovars is always required. Therefore, the aim of our study was study virulence genotyping of 28 Salmonella strains representing 9 Salmonella serovars (S.Agama, S.Blegdam, S. Enteritidis, S. Gueuletapee, S. Infantis, S. Kentucky, S. Montevideo, S. Typhimurium and S. Virchow) by Multiplex PCR technique targeting 17 virulence genes. Moreover, the resistance profile of those Salmonella strains to 15 antimicrobials was performed.
Materials and methods
Bacterial strains
Twenty eight Salmonella strains representing different Salmonella serovars (S.Agama, S.Blegdam, S.Enteritidis, S.Gueuletapee, S.Infantis, S.Kentucky, S.Montevideo, S.Typhimurium and S.Virchow) were used in this study. These Salmonella strains were previously isolated, purified, and identified in our laboratory from sick chickens suspected to infect with Salmonellosis.
Antibiotic sensitivity assay
Antimicrobial susceptibility testing to Salmonella strains belong to different serovars against different antimicrobials (Table 1) were determined by disk diffusion method (Bauer et al., 1966). Briefly, adjustment of bacterial inoculums to the 0.5 McFarland standard, streaking onto Mueller-Hinton agar plates, placing standard antibiotic disks (HIMEDIA®), and aerobic incubation at 37°C for 24 h were subsequently performed. The diameter of inhibition zone were measured and Salmonella strains were categorized to resistant, intermediate, or susceptible to different antimicrobials according to the CLSI guidelines (CLSI, 2017).
Polymerase chain reaction (PCR)
Bacterial DNA extraction and multiplex PCR amplification: Extraction of bacterial DNA was done for each Salmonella strain according to extraction kit instructions (GF-1 bacterial DNA extraction kit, vivantis, Malaysia). All bacterial DNA were kept in -20. Primers used in this study are listed in (Table 2). Three sets of multiplex PCR were accomplished for each sample to amplify different virulence genes (Skyberg et al., 2006) as follow: (set 1) amplified spvB, spiA, pagC, cdtB, and msgA. While (set 2) amplified invA, sipB, prgH, spaN, orgA, and tolC. Finally (set 3) amplified iroN, sitC, lpfC, sifA, sopB, and pefA. Amplification was performed in a 50 µl reaction mixture that included 1 µl of template DNA, 25 µl of master mix (Cosmo PCR Master Mix, UK), 2.5 µl of 50 mM MgCl2, 0.5 µl of 10 µM forward and reverse primers, and the reaction mixture was completed to 50 µl using dd H2O. Twenty five amplification cycles were run after 5 min at 95 C as follow: 30 sec at 94 C, 30 sec at 66.5 C, and 2 min at 72 C, with a final cycle of 10 min at 72 C, followed by a hold at 4 C. PCR products obtained were subjected to horizontal gel electrophoresis in 1.5% agarose, and the size of the amplicons was determined by comparison with DNA marker (VC 100bp Plus DNA Ladder, vivantis).
Statistical analysis
Antimicrobial resistance rates were analyzed using the chi-square test and GraphPad Prism 5.
Table 3: Antimicrobial resistance profiles of isolated Salmonella serovars
| Serovars (n) | Antimicrobial resistance | ||||||||||||||
|
AMCa |
AMPa |
CF | CPM | CTX | C | CIP | COT | DO | ETP | TE | NX | GEN | K | S | |
|
S. Blegdam (11) |
7 | 6 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
S. Typhimurium (7) |
7 | 6 | 2 | 4 | 4 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 2 | 2 |
|
S. Montevideo (3) |
3 | 3 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
|
S. Gueuletapee (1) |
1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
|
S. Agama (2) |
2 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
S. Enteritidis (1) |
1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
S. Kentucky (1) |
1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
S. Infantis (1) |
1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
S. Virchow (1) |
1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
0
|
| Total (28) | 24 | 22 | 9 | 5 | 8 | 2 | 1 | 2 | 3 | 0 | 6 | 1 | 0 | 4 | 2 |
| *Resistant % | 85.7% | 78.5% |
32 % |
17.8% |
28.5 % |
7% |
3.5 % |
7% | 10.7% | 0% |
21.3 % |
3.5 % |
0% |
14.3 % |
7% |
| *Intermediate % | 7% | 3.6% | 7% | 21.4% |
3.5 % |
10.7 % |
0% | 0% | 10.7% |
3.5 % |
25 % |
0% |
3.5 % |
17.8 % |
21.4 % |
| *Sensitive % | 7% | 17.8% |
60.7 % |
60.7% |
67.8 % |
82 % |
96.4 % |
92.8 % |
78.5% |
96.4 % |
53.6 % |
96.4 % |
96.4 % |
67.8 % |
71.4 % |
AMC Amoxicillin-clavulanic acid, AMP Ampicillin, CF Cefaclor, CPM Cefeprime, CTX Cefotaxime, C Chloramphenicol, CIP Ciprofloxacin, COT Trimethoprim sulfamethoxazole, DO Doxycycline, ETP Ertapenem, TE Tetracycline, NX Norfloxacin, GEN Gentamicin, K Kanamycin and S Streptomycin.
*The percentage of the total number of isolates resistant, intermediate, or susceptible for a particular antimicrobial is indicated in the last three rows below each antimicrobial.
a The significantly higher antimicrobial resistance rate.
Table 4: Multiple antimicrobial resistance patterns of Salmonella serovars
|
Multi-drug resistant antimicrobials |
Number of resistant Salmonella serovars |
Total (26) |
||||||||
|
S. Agama (n=2) |
S. Blegdam (n=11) |
S. Enteritidis (n=1) |
S. Gueuletapee (n=1) |
S. Infantis (n=1) |
S. Kentucky (n=1) |
S. Monte video (n=3) |
S. Typhim urium (n=7) |
S. Virchow (n=1)
|
||
| AMC, AMP | 1 | 6 | - | - | - | 1 | - | 1 | - | 9 |
| AMC,AMP,CF | - | 3 | - | - | - | - | 1 | - | - | 4 |
| AMC,AMP,CPM | - | - | - | - | - | - | - | 1 | - | 1 |
| AMC,AMP,CTX | - | - | - | - | - | - | 1 | 1 | - | 2 |
| AMC,AMP,COT | - | 1 | - | - | - | - | - | - | - | 1 |
| AMC,AMP,CF,TE | - | - | - | - | 1 | - | - | - | - | 1 |
| AMC,AMP,CF,CTX | - | - | 1 | - | - | - | - | - | - | 1 |
| AMC,AMP,CPM,CTX | - | - | - | - | - | - | - | 1 | - | 1 |
| AMC,AMP,DO,K | - | - | - | 1 | - | - | - | - | - | 1 |
| AMC,AMP,CF,C,TE | - | - | - | - | - | - | - | - | 1 | 1 |
| AMC,AMP,CTX,DO,TE | 1 | - | - | - | - | - | - | - | - | 1 |
AMC,AMP,CF,CPM, CTX,TE,K |
- | - | - | - | - | - | 1 | - | - | 1 |
AMC,AMP,CF,CPM, CTX,DO,TE,K,S |
- | - | - | - | - | - | - | 1 | - | 1 |
AMC,AMP,CF,CPM, CTX,C,CIP,COT,TE,NX,K,S |
- | - | - | - | - | - | - | 1 | - |
1 |
AMC Amoxicillin-clavulanic acid, AMP Ampicillin, CF Cefaclor, CPM Cefeprime, CTX Cefotaxime, C Chloramphenicol, CIP Ciprofloxacin, COT Trimethoprim sulfamethoxazole, DO Doxycycline, TE Tetracycline, NX Norfloxacin, K Kanamycin and S Streptomycin.
Table 5: Virulence genes percent detected in different Salmonella serovars
|
Virulence genes |
S. Agama (n=2) |
S. Blegdam (n=11) |
S. Enteritidis (n=1) |
S. Gueuletapee (n=1) |
S. Infantis (n=1) |
S. Kentucky (n=1) |
S. Montevideo (n=3) |
S. Typhimurium (n=7) |
S. Virchow (n=1)
|
Total percent |
| spvB | 2 | 10 | 1 | 0 | 0 | 0 | 0 | 6 | 1 | 71% |
| spiA | 2 | 11 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 100% |
| pagC | 2 | 10 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 96% |
| cdtB | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7% |
| msgA | 2 | 11 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 100% |
| invA | 2 | 11 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 100% |
| sipB | 2 | 7 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 86% |
| prgH | 2 | 11 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 100% |
| spaN | 2 | 4 | 1 | 0 | 1 | 1 | 3 | 5 | 0 |
61% |
| orgA | 1 | 4 | 1 | 0 | 1 | 1 | 3 | 5 | 0 | 57% |
| tolC | 2 | 11 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 100% |
| iroN | 2 | 4 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 50% |
| sitC | 2 | 10 | 1 | 1 | 1 | 1 | 2 | 7 | 1 | 93% |
| lpfC | 2 | 11 | 1 | 1 | 1 | 1 | 3 | 7 | 1 |
100% |
| sifA | 2 | 11 | 0 | 1 | 0 | 1 | 2 | 7 | 1 | 89% |
| sopB | 2 | 11 | 1 | 1 | 1 | 1 | 3 | 7 | 1 | 100% |
| pefA | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
21% |
Results
Antimicrobial resistance profile
Antimicrobial resistance profile to 28 Salmonella strains representing 9 serovars (S.Agama, S.Blegdam, S.Enteritidis, S.Gueuletapee, S.Infantis, S.Kentucky, S.Montevideo, S.Typhimurium and S.Virchow) were performed in this study. As shown in Table 3, the significantly higher rate of resistance was detected against amoxicillin-clavulanic acid (AMC) and ampicillin (AMP) (85.7% and 78.5% respectively) comparing to the significantly lower rate of resistance detected against etapenem (ETP), gentamicin (GEN), ciprofloxacin (CIP), and norfloxacin (NX) (0 %, 0%, 3.5%, and 3.5% respectively). The significantly high resistance rate to cephalosporin antibiotics were detected as follow: cefaclor (CF) (32%), cefotaxime (CTX) (28.5%), and Cefeprime (CPM) (17.8%) comparing to low resistance rate against ETP, GEN, CIP, and NX. Resistance to tetracycline (TE), kanamycin (K), trimethoprim/sulphamethoxazole (COT), chloramphenicol (C), and streptomycin (S) antimicrobials was 21.3%, 14.3%, 7% 7%, and 7% respectively. Multiple antimicrobial resistance was detected against 26 strains (Table 4): resistance to 2 out of 15 antimicrobials in 9 strains, resistance to 3 out of 15 antimicrobials in 8 strains, resistance to 4 out of 15 antimicrobials in 4 strains, and resistance to more than 4 antimicrobials in 5 strains. Interestingly, two S. Typhimurium strains were resist to 9 and 12 out of 15 antimicrobials used in this study.
Virulence genotyping
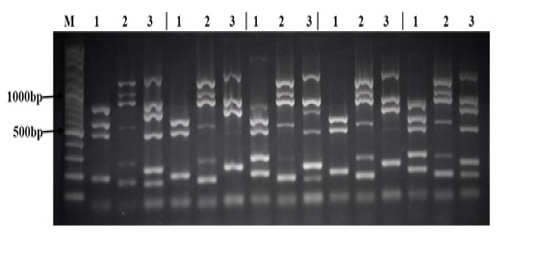
All 28 Salmonella strains were subjected to multiplex PCR targeting 17 virulence genes (spvB, spiA, pagC, cdtB, msgA, invA, sipB, prgH, span, orgA, tolC, iroN, sitC, lpfC, sifA, sopB, and pefA). As shown in Table 5, we detected 7 virulence genes in all Salmonella strains tested in this study (spiA, msgA, invA, prgH, tolC, lpfC, and sopB). The lowest rate of detection was cdtB and pefA genes (7% and 21% respectively). While other virulence genes were detected in different rate between different Salmonella strains as follow: spvB (71%), pagC (96%), sipB (86%), spaN (61%), orgA (57%), iroN (50%), sitC (93%), and sifA (89%). Interestingly, all 17 virulence genes were detected in S. Infants and one strain of S. Agama as shown in Figure (1).
There sets of multiplex PCR (1, 2, 3) for each isolate was performed as follow: Lane (1) is the result of the PCR reaction amplifying (from top to bottom) spvB, spiA, pagC, cdtB, and msgA. Lane (2) is the result of the PCR reaction amplifying (from top to bottom) invA, sipB, prgH, spaN, orgA, and tolC. Lane (3) is the result of the PCR reaction amplifying (from top to bottom) iroN, sitC, lpfC, sifA, sopB, and pefA. Lane (M) contains 100bp DNA Marker.
Discussion
A total of 28 Salmonella strains which representing 9 Salmonella serovars (S.Agama, S.Blegdam, S.Enteritidis, S.Gueuletapee, S.Infantis, S.Kentucky, S.Montevideo, S.Typhimurium and S.Virchow) were used in this study to perform antimicrobial resistance profile and virulence genotyping. In our study, a high resistance rates among different Salmonella strains were detected in AMC and AMP indicating the limited therapeutic value of these antibiotics to control salmonellosis. Salmonellae resistance to beta-lactam was previously reported in Egypt (Ammar et al., 2016; Khairy, 2015), Turkey (Siriken et al., 2015), Pakistan (Shah and Korejo, 2012), Brazil (Oliveira et al., 2006). Moreover, the resistance rates of Salmonella spp. to cephalosporin antibiotics, CF (2nd generation), CTX (3rd generation), and CPM (4th generation), were detected as 32%, 28.5%, and 17.8% respectively. Development of resistance against cephalosporins was previously detected (Abo-Amer and Shobrak, 2015; Elkenany et al., 2019; Mir et al., 2015) which has a public health consequences as these antimicrobials are used to treat serious Salmonella infections in human. Unfortunately, the resistance rate of Salmonella strains tested in this study to 3 or more antimicrobials was 65%. Interestingly, two S. Typhimurium strains were resist to 9-12 out of 15 antimicrobials. Thus, increasing resistance rates and multiple antimicrobial resistance among Salmonella strains (Ammar et al., 2016; Elkenany et al., 2019; Yu et al., 2021) could be due to haphazard use of antimicrobials at recommended doses or at sub-therapeutic doses which representing a public health hazard. Antimicrobial resistance among Salmonella is a serious public health problem that needs to be monitored continuously. Furthermore, using of alternatives instead of antibiotics to control salmonellosis in poultry is required.
We performed a multiplex PCR targeting 17 virulence genes of Salmonellae related to adherence, invasiveness, entry to non-phagocytic cells and killing of macrophages, survival within macrophage, growing within the host, iron acquisition, and toxin biosynthesis. Genes required for host recognition and invasion (invA, prgH, tolC, lpfC, and sopB) and also required for survival of Salmonella within macrophages (spiA and msgA) were detected in all Salmonella strains tested in this study. Both cdtB and pefA virulence genes were rarely detected (7% and 21% respectively) which agree with results previously reported (Skyberg et al., 2006). Moreover, cdtB gene which responsible for toxin biosynthesis (Haghjoo and Galan, 2004) was only detected in both strains of S. Agama while PefA gene encoded by virulence plasmid was detected in S. Agama (2/2) and S. Typhimurium (4/7). Interestingly, all virulence genes were detected in S. Infants and 1 out of 2 S. Agama tested in this study. S. Agama was previously isolated from the poultry environment, dead birds, and apparently healthy birds in Nigeria (Ahmed et al., 2019). Furthermore, it is a zoonotic pathogen as it was a cause of diarrhea (Bélard et al., 2007; Kudaka et al., 2006), neonatal meningitis and septicemia (Heaton et al., 2015).
Conclusion
Antimicrobial resistance profile and virulence genotyping to Salmonella strains representing 9 Salmonella serovars previously isolated and identified in our laboratory were performed in this study. Multidrug resistant Salmonella strains were described and many virulence genes were detected among Salmonella strains. Finally, continuous monitoring of antimicrobial resistant Salmonella strains, using of alternatives instead of antimicrobials in poultry, and strict public health and food safety regimens are required to decrease the human health risk associated with Salmonellosis.
Acknowledgment
We would like to thank Prof. Dr. Ebtehal Abd El Aty Elsayed, chief researcher in microbiology and serology unit in Animal Health Research Institute, for their technical support. This work was funded by national project (12010140) under a title (control of salmonellosis in chickens) in National Research Centre. Furthermore, the project has been approved by Medical Research Ethic Committee in National Research Centre under number 19157.
Conflict of interest
The authors declare that they have no conflict of interest.
authors contribution
Nagwa S. Rabie and Samy, A.A. carried out the research design and revised the manuscript. Abdelbaki, M.M., Hanaa S. Fedawy, M. A. Bosila, Dalia M. Sedeek, and Aly M. Ghetas performed antimicrobial resistance profile. Hanaa S. Fedawy also participted in revision of the maniscript. Aly M. Ghetas and Abdelbaki, M.M. achieved virulence genotyping. Aly M. Ghetas analyzed data and wrote the manuscript.
References