Advances in Animal and Veterinary Sciences
Research Article
Inhibitory Effects of Carvacrol on BlaTEM and Exos Genes Expression in ESβL Producing Pseudomonas aeruginosa Isolated from Kidney Lesions of Broiler Chickens
Ismail A. Radwan1, Salama A.S. Shany2, Sara S.E. Amin1, Ahmed H. Abed1*
1Bacteriology, Mycology and Immunology Department, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef 62511, Egypt; 2Poultry Diseases Department, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef 62511, Egypt.
Abstract | Pseudomonas aeruginosa is an opportunistic environmental pathogen causing serious problems in poultry farms. Our study investigated the prevalence of P. aeruginosa in 80 examined kidney samples of broiler chicken where 6 P. aeruginosa isolates (7.5%) were isolated. The extended spectrum β-lactamases (ESβLs) production was detected phenotypically in P. aeruginosa isolates using modified CLSI ESβLs confirmatory test and 66.7% of isolates were ESβLs producers. PCR was conducted on all isolates for detection for blaTEM and exoS genes that were found in 66.7 and 100% of isolates, respectively. Antibacterial activity of carvacrol oil was tested against all P. aeruginosa isolates at concentrations of 800, 400, 200, 100 and 50 µl/ml. Concentrations of 800, 400 and 200µl/ml showed complete growth inhibition while 100 and 50 µl/ml concentrations inhibited the growth of 66.7% and 33.3% of isolates, respectively. Real time-PCR (RT-PCR) was conducted on the non-inhibited P. aeruginosa isolates at 100 µl/ml concentration after treatment with carvacrol oil for detection of the possible effect on blaTEM and exoS genes expression and the results indicated mild reduction of both genes expression after treatment (0.6-0.8 folds). It was concluded that carvacrol oil could be used as alternatives for synthetic antimicrobial drugs.
Keywords | P. aeruginosa, ESβL, blaTEM gene, exoS gene, Carvacrol, Broiler chickens, Kidney lesions
Received | February 16, 2021; Accepted | May 09, 2021; Published | July 28, 2021
*Correspondence | Ahmed H. Abed, Bacteriology, Mycology and Immunology Department, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef 62511, Egypt; Email: [email protected], [email protected]
Citation | Radwan IA, Shany SAS, Amin SSE, Abed AH (2021). Inhibitory effects of carvacrol on BlaTEM and Exos genes expression in ESβL producing Pseudomonas aeruginosa isolated from kidney lesions of broiler chickens. Adv. Anim. Vet. Sci. 9(9): 1408-1415.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.9.1408.1415
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Lukkananukool et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Pseudomonas is a good example of environment-associated infections that causing serious problems in poultry industry because epidemics may spread rapidly through poultry flocks causing mortality in all ages (Satish and Priti, 2015). P. aeruginosa is the most common opportunistic ubiquitous pathogen that often exists in decaying vegetation, soil and water as well as other humid environments (Bakheet and Torra, 2020). P. aeruginosa is a serious avian pathogen as well as zoonotic bacterial agent that can cause nosocomial infections (El-Sayed et al., 2016). The infection may take place via skin wounds, contaminated vaccines, egg inoculation or dipping or via contamination of needles used for injection, and infection can spread between flocks on the same premises under inadequate hygienic conditions (Mohamed, 2004). It can cause respiratory affections, septicemia and other forms in birds (Bakheet and Torra, 2020) inducing a significant economic losses due to high mortality (Elsayed et al., 2016) and can be highly virulent causing 50-100% mortality in experimentally inoculated 4 weeks old chickens (Kebede, 2010).
Pseudomonas species have been reported as a cause of bacterial nephritis (Lierz, 2003) which usually occurs after systemic infection and bacteria reach the kidneys through the renal arteries or the renal portal system while ascending infection through the ureters rarely occurs (Ameen et al., 2015).
A great problem of P. aeruginosa is its resistance to various antibacterial agents and even newly developed antibiotics have failed to reduce the mortality rate associated with its infection (Ali et al., 2009). The extended spectrum β-lactamases (ESβLs) and plasmid-mediated ampC β-lactamase are essential causes behind the antimicrobial resistance (Carmo et al., 2014). ESβLs production represents high risk in the treatment of pseudomonal infection. ESβLs are widely reported all over the world and have been associated with successful enterobacterial clones having huge epidemic potential (Zahar et al., 2009). ESβLs are plasmid-mediated β-lactamase enzymes that hydrolyze penicillins, narrow spectrum β-lactamas, 3rd and 4th generation cephalosporins and monobactams, meanwhile β-lactamase inhibitors; as clavulanic acid, can inhibit them (Poulou et al., 2014). ESβLs coding plasmids may carry also additional β-lactamase genes and other resistance genes for other antimicrobial classes (Carattoli, 2009). This can restrict the treatment options for ESβL-producing pathogens and enhance the intra- and inter-species spreading of ESβLs (Zahar et al., 2009). Therefore, phenotypic detection of ESβLs within bacteria is essential for epidemiological purposes and restriction of the dissemination of resistance mechanisms. The Clinical and Laboratory Standards Institute recommended a phenotypic confirmatory combined-disc test for ESβL production (CLSI, 2015). It was based on detection of the growth-inhibition zones around both cefotaxime and ceftazidime discs with or without clavulanate (CA). Stuart et al. (2011) have also proposed different combined-disc and double-disc synergy tests depending on the synergy of CA with different expanded spectrum cephalosporins and aztreonam. There are many produced groups of ESβLs but the most commonly existed in clinical Gram-negative isolates are SHV, CTX-M and TEM enzyme types (Bush and Fisher, 2011).
P. aeruginosa exoenzyme S (ExoS) is a type III secretion (TTS) effector which has been considered an antiphagocytic virulence factor of P. aeruginosa that enabling it to overcome the host defense mechanism leading to the establishment of infection and tissue damage (Rocha et al., 2003). It catalyzes the transfer of the ADP ribose moiety of NAD+ to many eukaryotic cellular proteins, but its preferred substrates are a subset of the small GTP-binding proteins of 21-25 kDa (Barbieri and Sun, 2004).
The spread of drug resistant strains of microorganisms necessitates the discovery of new classes of antibacterial and compounds that inhibits these resistance mechanisms. Bacterial resistance modulators can enhance the activity of an antimicrobial agent against a resistant strain. Such compounds may target a resistance mechanism such as the inhibition of multidrug resistance (MDR) e.g. inhibition of the NorA efflux mechanism or act in a synergistic fashion via an uncharacterized mechanism (Gibbons, 2005). The essential oils (EOs) from many plants are known to possess antimicrobial activity (Radwan et al., 2016). Oregano oil and its major phenolic components carvacrol oil have a wide spectrum of antimicrobial activity (Nostro et al., 2007).
The present study was conducted to detect the possible effects of carvacrol oil on blaTEM and exoS genes expression of P. aeruginosa recovered from kidneys of broiler chickens to overcome the drug resistant isolates.
MATERIALS AND METHODS
Ethical approval
This study was approved from Beni-Suef University, Institutional Animal Care and Use Committee (BSU-IACU/ http://www.bsu.edu.eg).
Samples
Eighty kidney samples were collected from diseased broiler chickens aged from 2-4 weeks from different farms in Beni-Suef and El-Fayoum Governorates during the period from January 2019 up to October 2019. These chickens were subjected to postmortem examinations and showed gross pathological kidney lesions. All samples were collected in sterile containers and transferred rapidly in ice box to the laboratory of Bacteriology, Mycology and Immunology Department, Faculty of Veterinary Medicine, Beni-Suef University.
Isolation and identification of P. aeruginosa isolates
The collected kidney samples were inoculated under aseptic conditions into tryptone soya broth (Oxoid) and incubated aerobically at 37˚C for 18-24 h. A loopful of broth culture was cultivated onto cetrimide agar and incubated aerobically at 37˚C for 24-72 h. All the recovered isolates were identified morphologically; using Gram’s stain, and biochemically according to schemes described by Quinn et al. (2002). The following tests were used; oxidase, catalase, indole, methyl red, Voges Proskauer, citrate utilization, urease, TSI and sugar oxidation; especially glucose, mannitol and mannose. Other characteristics including pigment production, motility test haemolysis onto blood, growth at 4 and 42ºC and agar were included.
Moreover, The Vitek 2 compact system using ID-GN kits was applied on pure cultures for confirmative identification of P. aeruginosa isolates according to BioMérieux (2013).
Phenotypic detection of EsβLS producing isolates (CLSI, 2015)
ESβL production was tested with the CLSI confirmatory test using modified double disc diffusion test by using cefotaxime (CTX, 30 µg), ceftazidime (CAZ, 30 µg), cefepime (FEP, 30 µg) and aztreonam (ATM, 30 µg) discs alone and in combination with amoxicillin-clavulanic (AMC, 30 µg) (Oxoid, Basing Stoke, UK). The discs of CTX, CAZ, FEP and ATM were manually placed around AMC disc with 20 mm center to center and incubated at 37°C for 8 h. The test was regarded as positive when increasing the zone of growth-inhibition around one or more of discs with AMC to 5 mm or more than the diameter around the disc containing them alone.
PCR for detection of BLATEM and EXOS genes in P. aeruginosa isolates
PCR was applied on all P. aeruginosa isolates for detection of blaTEM and exoS genes. Genomic DNA was extracted by QIAamp® DNA extraction Mini Kit (Cat. No. 51304 supplied from QIAGEN, USA), according to manufacturer’s instructions. Extracted DNA was kept at -80°C until used in PCR amplification. Oligonucleotide primers sequences as well as amplified products for the targeted genes were illustrated in Table 1. The reaction was performed in a volume of 25 µl consisting of 12.5 µl of 2X PCR master mix, 1µl of each 20 pmol primers, 6µl of DNA extract, and the volume was completed to 25µl using sterile deionized water. The temperature and time conditions of the primers during PCR were shown in Table 2.
Detection of minimum inhibitory concentration (MIC) of carvacrol oil against P. aeruginosa isolates using agar dilution method
Different concentrations (800, 400, 200, 100, 50 µl/ml) of carvacrol (Sigma Aldrich, Germany) were prepared and tested for their antibacterial activity against all P. aeruginosa isolates using agar dilution method according to Radwan et al. (2018). Briefly, the tested isolates were grown on tryptone soya agar (TSA) at 37˚C for 24 h, then cells were suspended in physiological saline adjusting the concentration to 1×108 CFU with (equivalent to McFarland standard tube 0.5). TSA was prepared and autoclaved at 121˚C for 15 min. and kept at 55˚C and then the tested oils were dissolved in Dimethyl Sulpho Oxide (DMSO) with a ratio 1:9, sterilized by filtration (pore size 0.45 µm), and mixed with TSA according to the tested concentrations. The oil-agar medium (10 ml) was poured into sterile petri dishes and was solidified. Then, equal amounts of the bacterial suspensions were inoculated and speared onto the agar plates and incubated at 37˚C for 24-48 h.
The maximum concentration of carvacrol that did not completely inhibit the bacterial growth was selected for detection of the possible effect on blaTEM and exoS gene expression.
Sybr green RT-PCR for BLATEM and EXOS genes before and after carvacrol treatment
SYBR Green RT-PCR was applied for detection of fold changes of blaTEM and exoS genes before and after carvacrol treatment. RNA was extracted by RNeasy Mini Kit (Qiagen Cat. No.74104) according to manufacturer’s instructions. Oligonucleotide primers and probes used in SYBR Green RT-PCR illustrated in Table 1 beside Primers targeting Pseudomonas16S rDNA primers (F: 5’-GACGGGTGAGTAATGCCTA-3’ and R: 5’-CACTGGTGTTCCTTCCTATA-3’ according to Spilker et al. (2004) were used.
The reaction was performed in a volume of 25 µl consisting of 12.5 µl of 2X SYBR Green PCR master mix, 0.25 μl reverse transcriptase, 0.5µl of each 20 pmol primers, 3µl of RNA extract, and the volume was completed to 25µl using sterile RNase free water. The temperature and time conditions of the primers during SYBR green RT-PCR were shown in Table 3. Analysis of the SYBR green RT-PCR results was according to the “ΔΔCt” method stated by Yuan et al. (2006).
Table 1: Primers of blaTEM and exoS genes used in cPCR and SYBR Green RT-PCR.
| Primer | Amplified product | Primer sequence (5'-3') | Reference | |
|
blaTEM |
516 bp |
ATCAGCAATAAACCAGC CCCCGAAGAACGTTTTC |
F R |
Colom et al. (2003) |
|
exoS |
118 bp | GCGAGGTCAGCAGAGTATCG TTCGGCGTCACTGTGGATGC |
F R |
Winstanley et al. (2005) |
Table 2: Cycling conditions of the different primers during PCR.
| Gene | Primary denaturing | Secondary denaturing | Annealing | Extension | No. of cycles | Final extension |
|
blaTEM |
94˚C/5 min |
94˚C/30sec. |
54˚C/40sec. |
72˚C/45sec. |
35cycles |
72˚C/10min. |
|
exoS |
94˚C/5min. |
94˚C/30sec. |
55˚C/30sec. |
72˚C/30sec. |
35cycles |
72˚C/7min. |
Table 3: Cycling conditions for SYBR green RT-PCR.
| Target gene | Reverse transcription | Primary denatu ration | Amplification (40 cycles) | Dissociation curve (1 cycle) | ||||
| Secondary denaturation | Annealing (Optics on) | Extension | Secondary denaturation | Annealing | Final denat uration |
|||
| Pseudomonas 16S rDNA |
50˚C/ 30min |
94˚C/ 15min |
94˚C/15sec. |
50˚C/40sec. |
72˚C/ 40sec |
94˚C/1min
|
54˚C/1min |
94˚C /1min
|
|
blaTEM |
54˚C/40sec. | 54˚C/1min | ||||||
|
exoS |
55˚C/30sec. | 55˚C/1min | ||||||
Table 4: RT-PCR for P. aeruginosa isolates before and after treatment with carvacrol oil.
| Sample No. | Type | Sample (ID) | 16S rDNA | blaTEM | exoS | ||
| CT | CT | Fold change | CT | Fold change | |||
| 1 | Control | 2 | 20.73 | 21.19 | - | 20.55 | - |
| Treated | 2A | 20.82 | 21.61 | 0. 7955 | 21.08 | 0.7371 | |
| 2 | Control | 3 | 20.51 | 22.05 | - | 21.10 | - |
| Treated | 3A | 20.74 | 22.91 | 0.6462 | 22.12 | 0.5783 | |
RESULTS AND DISCUSSION
Prevalence of ESβLS producing P. aeruginosa isolation from cases kidney lesions in broiler chickens
Out of 80 kidney samples from diseased broiler chickens, 6 P. aeruginosa isolates (7.5%) were recovered. Phenotypic detection of ESβLs producing P. aeruginosa isolates; using modified CLSI ESβLs confirmatory test, revealed that 4 isolates (66.7%) were ESβLs producers.
PCR OF P. aeruginosa isolates for detection of BLATEM and EXOS genes
PCR results revealed that out of 6 examined P. aeruginosa isolates, blaTEM was positive only in 4 isolates (66.7%) those were confirmed phenotypically as ESβLs producers while the other 2 negative ESβLs producers isolates didn`t exist blaTEM gene (Figure 1). Meanwhile, exoS gene was positive in all isolates (n=6, 100%) (Figure 2).
Detection of mic of carvacrol against P. aeruginosa isolates
Carvacrol oil completely inhibited the growth of the tested P. aeruginosa isolates at concentrations of 800, 400 and 200 µl/ml. Meanwhile, at concentrations of 100 and 50 µl/ml, the growth of 4 (66.7%) and 2 (33.3%) isolates were inhibited, respectively.
Sybr green RT-PCR for BLATEM AND EXOS genes in P. aeruginosa isolates before and after treatment with carvacrol
The quantitative RT-PCR (qRT-PCR) was applied on the two P. aeruginosa isolates; those not inhibited with carvacrol treatment at concentration of 100 µl/ml, for detection of possible fold changes of blaTEM and exoS genes expression after treatment with carvacrol. The results were illustrated in Table 4 and Figures 3, 4 and 5).
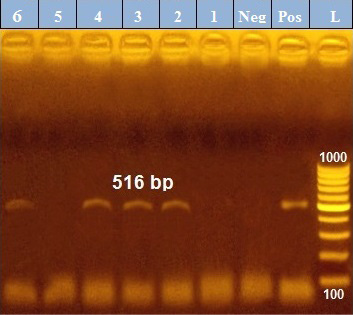
Figure 1: PCR amplification of the 516 bp fragment of blaTEM resistance gene from 6 P. aeruginosa isolates (1-6), Pos. (control positive), Neg. (control negative).
Regarding blaTEM gene, the results of qRT-PCR showed that the fold changes in the two P. aeruginosa isolates after treatment were 0.7955 and 0.6462 indicating mild reduction in the blaTEM gene expression after carvacrol treatment to 0.8 and 0.6 folds, respectively (Table 4 and Figure 4). Meanwhile, the fold changes of exoS gene after carvacrol treatment were 0.7371 and 0.5783, indicating also mild reduction in exoS gene expression to 0.7 and 0.6 folds, respectively (Table 4 and Figure 5).
Pseudomonas aeruginosa is a good example of ubiquitous opportunistic environmental pathogens; found in soil, water, feed and farm equipment, causing serious problems in poultry farms including respiratory infections, septicaemia and other forms when introduced into tissues of susceptible birds (Bakheet and Torra, 2020) resulting in significant economic losses due to high mortalities in all ages (Satish and Priti, 2015). Therefore, application of sanitary measures should be taken in considerations in the poultry husbandry especially feeds and water supply. Outbreaks of P. aeruginosa infection may cause mortality rate that reach 20-90% (Shukla and Mishra, 2015). High mortalities of P. aeruginosa infections are due to the existence of different virulence factors, innate and acquired MDR as well as immune compressed hosts (Poole, 2011).
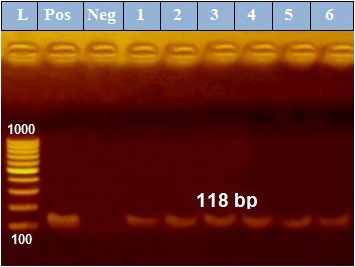
Figure 2: PCR amplification of the 118 bp fragment of exoS resistance gene from 6 P. aeruginosa isolates (1-6), Pos. (control positive), Neg. (control negative).
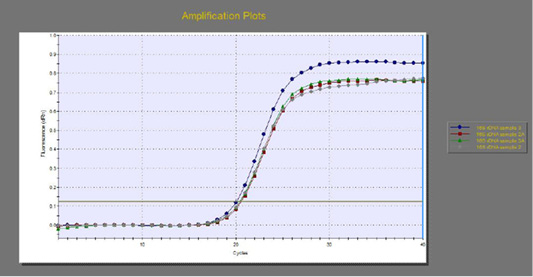
Figure 3: The qRT-PCR of 16s rDNA gene from 2 P. aeruginosa isolates before and after treatment with 100µl/ml carvacrol.
In this study, the prevalence of P. aeruginosa isolation from kidney lesions in broiler chickens was 7.5%. This result was supported with that obtained by Al-Hiyali et al. (2005) in Iraq, who studied 80 broiler chickens cases of damaged kidney and isolated P. aeruginosa from 14 cases with a percentage of 17.5%. Other studies reported P. aeruginosa as a cause of bacterial nephritis (Lierz, 2003; Satish and Priti, 2015). It can reach the kidney via circulation secondary to systemic disease and rarely can ascend the ureters (Ameen et al., 2015).
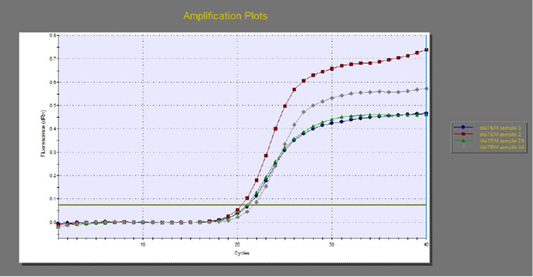
Figure 4: The qRT-PCR of blaTEM gene from 2 P. aeruginosa isolates before and after treatment with 100µl/ml carvacrol.
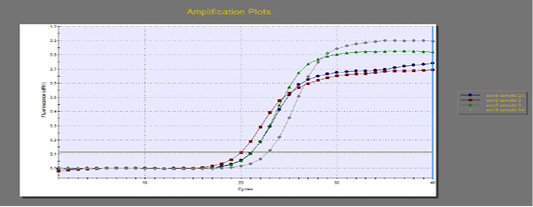
Figure 5: The qRT-PCR of exoS gene from 2 P. aeruginosa isolates before and after treatment with 100µl/ml carvacrol.
Treatment of infections caused by P. aeruginosa is difficult since it has high resistance to various antimicrobials (Ali et al., 2009). ESβLs are plasmid-mediated β-lactamases that hydrolyze cephalosporins and monobactams with an oxyimino side chain while they can be inhibited by β-lactamase inhibitors; as clavulanic acid, (Fisher, 2011). CLSI recommended a phenotypic confirmatory combined-disc test for detection of ESβL production (CLSI, 2015).
In our study, ESβLs producing P. aeruginosa isolates from kidney lesions were phenotypically detected using modified CLSI ESβLs confirmatory test and 66.7% of examined isolates were confirmed as ESβLs producers. Much lower results were previously recorded (Umadevi et al., 2011; 19.4%, (Rafiee et al., 2014); 6.5% of human isolates from burns, (Zafer et al., 2014); 7.4% among isolates from cancer patients; (Shaikh et al., 2015); 25.1% of isolates using double disc synergy test. Moreover, Begum et al. (2013) noticed ESβLs production in 35.4 % of the examined Pseudomonas aeruginosa isolates by double disc diffusion method using AMC with CAZ, CTX, ceftriaxone and ATM.
The emergence and spreading of β-lactam resistance in nosocomial P. aeruginosa and Enterobacteriaceae became a serious problem particularly the increased resistance against carbapenems and 3rd and 4th generation cephalosporins (Pfeifer et al., 2010). ESβLs are of the main causes of β-lactam resistance among Gram-negative bacteria (Rawat and Nair, 2010). P. aeruginosa pursue multiple molecular mechanisms for emerging the resistance to these antibiotics; (a) ESβL generation, (b) acquiring the ESβL encoding genes as SHV, CTX-M and TEM β-lactamases from environmental bacteria, (c) increasing the chromosome-encoded β-lactamase genes (bla) expression, (d) movement of bla genes by fusion with integrons and horizontal transferring into other Gram-negative bacteria, (e) spreading of plasmid-mediated carbapenemases (such as metallo-β-lactamases and KPC), (f) stop porin genes expression and/or efflux pump-based antibiotic resistance (Pfeifer et al., 2010).
ExoS is a virulence factor of P. aeruginosa having an antiphagocytic activity allowing the bacteria to overcome the host immunity and consequently establishment of infection and damage of tissues (Rocha et al., 2003). ExoS and other exotoxins share in tolerance of the innate immune responses and possess enzymatic activities that leading to disturbance in host cells physiology preventing bacterial clearance. Presence of substrate targets and co-factors in the eukaryotic cells are responsible for these effectors activation giving specificity to these toxins in the eukaryotic cells (Stato et al., 2006).
In this study, PCR was conducted on all P. aeruginosa isolates (n=6) for detection for blaTEM and exoS genes. Results revealed the existence of blaTEM in isolates confirmed phenotypically as ESβLs producers only (n=4; 66.7%) while others non- ESβLs producers were negative for blaTEM gene. These results were coincided with that considering ESβLs as of the major causes of β-lactam resistance among Gram-negative bacteria (Rawat and Nair, 2010) and that considering TEM enzyme type as of the most commonly existed ESβLs in clinical isolates (Bush and Fisher, 2011).
Meanwhile, exoS gene was existed in all isolates (100%). Different percentages of exoS gene were reported in previous studies; Zhao et al. (2012) detected exoS in 58% of examined P. aeruginosa isolates, Tartor and El-Naenaeey (2016) reported that 79% of P. aeruginosa isolates expressed exoS which was found in more virulent strains and Benie et al. (2017) detected exoS in 89% of P. aeruginosa isolates.
Since ancient times, the antimicrobial impact of Eos and their components extracted from aromatic and medicinal plants, both on health and food preservation have been recognized (Radwan et al., 2018). Moreover, EOs from many plants are known to possess antimicrobial activity (Radwan et al., 2016). The antimicrobial actions of EOs are associated with their hydrophobicity leading to increase the cell permeability and consequently leakage of cell components (Lambert et al., 2007). The concept of using a compound that inhibits resistance in a bacterium, which may be employed with a conventional antibiotic, was well proven (Gibbons, 2005).
In our study, carvacrol oil was tested for its antibacterial activity against P. aeruginosa isolates at concentrations of 800, 400, 200, 100 and 50 µl/ml for detection of MIC against P. aeruginosa isolates. Complete growth inhibition of the tested isolates was recorded at concentrations of 800, 400 and 200µl/ml. Therefore, the MIC of carvacrol against P. aeruginosa isolates was 200µl/ml. Meanwhile, 100 and 50µl/ml concentrations inhibited the growth of 66.7% and 33.3% of isolates, respectively. Such results supported those reported by Al-Sayed (2017) who studied the antibacterial effect of carvacrol oil on P. aeruginosa and recorded complete growth inhibition at concentration of 1% while concentration of 0.1% inhibited 73.3% of the tested isolates. On the same context, Radwan et al. (2016) studied the antibacterial effect of oreganium EO on E. coli and recorded complete bacterial growth inhibition at concentrations of 1% and 0.5%.
In the present study, the two P. aeruginosa isolates which not inhibited with treatment with carvacrol at concentration of 100 µl/ml was selected for detection of the possible effect on blaTEM and exoS gene expression using qRT-PCR after carvacrol treatment. The qRT-PCR results indicated mild reduction of the blaTEM and exoS genes expression after carvacrol treatment to 0.6-0.8 folds.
Carvacrol is the bioactive lipophilic and phenolic component; representing the major constituent of thyme and oregano EOs comprising 86.5 %, having high inhibitory activity against various pathogens (Bharti et al., 2013). The antibacterial activity of carvacrol was attributed to disintegration of the outer membrane of Gram-negative bacteria releasing LPS and enhancing the cytoplasmic membrane permeability to ATP (Burt, 2004). Carvacrol has been suggested to occupy more area than the typical space between the fatty acid chains of two adjacent phospholipid molecules interfering with the van der Waals interactions between the chains expanding the liposomal membrane using fluorescent probes and consequently affects fluidity (Ultee et al., 2002).
CONCLUSIONS AND RECOMMENDATIONS
Pseudomonas aeruginosa is an opportunistic environmental pathogen causing serious problems in poultry farms. The prevalence of P. aeruginosa in the examined kidney samples of broiler chickens was 7.5%. Most of isolates (66.7%) were ESβLs producer harboring both blaTEM and exoS genes. Carvacrol oil treatment showed an antimicrobial activity against the isolates with a MIC of 100µl/ml and induced mild reduction of the blaTEM and exoS genes expression after carvacrol treatment. Therefore, carvacrol and other EOs could be used as alternatives for synthetic antimicrobial drugs.
ACKNOWLEDGMENTS
The authors would like to thank the Bacteriology, Mycology and Immunology Department staff at the Faculty of Veterinary Medicine, Beni-Suef University for providing technical help.
Novelty Statement
The present work studied the antibacterial effect of carvacrol oil on ESβL Producing Pseudomonas aeruginosa isolated from kidney lesions of broiler chickens and detecting its inhibitory effects on the blaTEM and exoS genes expression in P. aeruginosa isolates.
AUTHOR’S CONTRIBUTION
Idea and Conceptualization, IAR and AHA; Sample collection, AHA, SSEA and SASS; Methodology and data analysis IAR, AHA, SASS and SSEA; Original draft preparation, AHA and SASS; Reviewing and editing, IAR, AHA, SASS and SSEA.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES






