Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (2): 98 – 103Molecular Characterization of Segment 6 of Bluetongue Serotype 16 of Sheep Origin from India
Koushlesh Ranjan, Gaya Prasad, Pawan Kumar, Prasad Minakshi*
*Corresponding author: [email protected]
ARTICLE CITATION:
Ranjan K, Prasad G, Kumar P, Minakshi P (2014). Molecular characterization of segment 6 of bluetongue serotype 16 of sheep origin from India. Adv. Anim. Vet. Sci. 2(2): 98 – 103.
Received: 2013–12–12, Revised: 2013–12–21, Accepted: 2013–12–22
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.2.98.103
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
The bluetongue (BT) is a Culicoides vector transmitted viral disease of ruminant. The disease is endemic in India and many other countries of the world. In the present study, the bluetongue virus (BTV) isolate BT16/07, originally isolated from sheep, was cultivated in BHK–21 cell line and showed the characteristic cytopathic effect (CPE) within 48 hours. The viral nucleic acid (dsRNA) was extracted using Trizol method and tested in RNA–poly acrylamide gel electrophoresis (RNA–PAGE). The RNA–PAGE analysis of viral nucleic acid revealed a BTV–characteristic 3:3:3:1 migration pattern. The cDNA prepared from viral nucleic acid was subjected to non–structural gene 1 (NS1) based group–specific PCR to confirm the genetic nature of the virus. The group–specific NS1 gene based PCR showed a single 366bp amplicon and confirmed the isolate as BTV. The serotype specific PCR was conducted using VP5 gene specific primers for serotyping of the BT16/07 isolate. The serotype–specific PCR, using BTV16 VP5 gene–specific primer, showed a single amplicon of 757bp. The remaining BTV serotype–specific primers fail to amplify the respective gene sequence. The VP5 gene–specific PCR amplicon was cloned and subsequently sequenced. The nucleotide sequence obtained was subjected to in–silico restriction enzyme analysis and phylogenetic analysis. The phylogenetic tree, sequence identity analysis and in–silico restriction enzyme analysis (REA) indicate the eastern origin of the BT16/07 isolate.
INTRODUCTION
Bluetongue (BT) is an economically important arthropod–transmitted viral disease of wild and domestic ruminants. The disease is caused by bluetongue virus (BTV), a member of the genus Orbivirus and family Reoviridae (Murphy et al., 1995). The BTV is transmitted between vertebrate hosts by Culicoides (biting midges) vector (Mellor, 1990). The outer capsid of viral particle is comprised of two proteins, VP2 and VP5, where VP5 is serotype specific next to the major VP2 protein. The inner core of viral particle is consists of two major proteins, VP7 and VP3, three minor proteins viz. VP1, VP4, VP6, and ten segments of dsRNA genome. In addition to above structural proteins, there are non–structural proteins i.e. NS1, NS2, NS3/NS3a and NS4, expressed in virus infected host cells (Mertens et al., 1989; Ratinier et al., 2011; Belhouchet et al., 2011). The BT disease is characterised by severe clinical signs such as high fever, lameness, swelling of lips and tongue, abortion and stillbirth. The more severe forms of the disease are frequently seen in sheep and in white–tailed deer (Darpel et al., 2007; Howerth et al., 1989). Buffalo, cattle and goats act as silent reservoirs and remaining viraemic for several months (Maclachlan et al., 2009). There are 24 distinct serotypes of BTV have been reported from different parts of the world (Mertens et al., 2004). However, two more serotypes i.e. BTV25 from Switzerland and BTV26 from Kuwait have been isolated recently (Hofmann et al., 2008; Maan et al., 2011). BTV is endemic in India and so far 21 different serotypes have been reported from different geographical regions based upon serum neutralization and virus isolation (Prasad et al., 2007). Recently 22nd serotype i.e. BTV 21 has been reported from Andhra Pradesh state of the country (Susmitha et al., 2012). Recently, new BTV isolate has been isolated and serotyped as BTV 10 from India (Gollapalli et al., 2012; Maan et al., 2012a). The present study was carried out to characterize the VP5 gene of BTV/16/07, to study of genetic variation and to determine the phylogenetic relationship of this Indian isolate with isolates reported previously.
MATERIALS AND METHODS
Sample Origin
The virus isolat,e was obtained from Andhra Pradesh state in 2007. The sample was designated as isolate BT16/07. The virus was originally isolated from blood sample of Nellore breed of sheep having high temperature (40ºC) and nasal–oral lesions.
Cultivation of Virus and Isolation of Viral Nucleic Acid (dsRNA)
The virus sample was originally propagated in 9–11 day old embryonated chicken egg and finally cultivated in BHK–21 cell line monolayer. The virus sample was further passaged for 10 passages in BHK–21 cell line.
After appearance of about 75% cytopathic effect (CPE), the BHK–21 cells along with virus were centrifuged at 5000rpm in a table top centrifuge (REMI, India) for 5 minute. The supernant was discarded and the pelleted material was used for extraction of viral dsRNA using TRIzol (Life technologies, USA) method as per manufacturer’s protocol.
RNA–Polyacrylamide Gel Electrophoresis (RNA–PAGE)
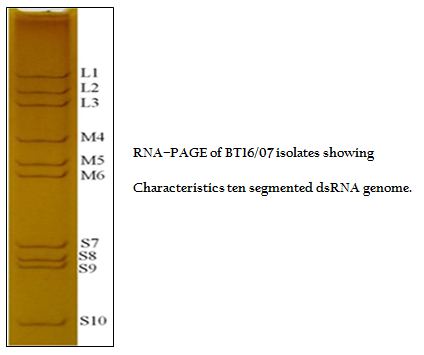
The viral nucleic acid (dsRNA) was analyzed by RNA–PAGE using 8% resolving gel in discontinuous buffer system (Laemmli, 1970). The gel was stained with silver nitrate to visualize the 10 dsRNA segments for confirmation of sample as BTV (Svensson et al., 1981).
Serogroup and Serotype Specific Primers
The serogroup–specific NS1 gene based PCR was performed for confirmation of the isolate as BTV using published primers (Kovi et al., 2005). Additionally, the BTV serotype 16 VP5 gene specific primer pairs P1 (776–795nt) – 5’–AGAGCGATCGAGGGCGCGTA–3’ and P2 (1532–1513nt) – 5’–TTGCGCGCCGTTGGAAATGC–3’ were designed using nucleotide sequences of BTV16 available in GenBank using Primer Blast tool (http://www.ncbi.nlm.nih.gov/tools/primer–blast/). The expected PCR amplicon size of primer pair, P1 and P2 was 757 bp.
PCR Amplification of VP5 and NS1 Gene
Viral genomic dsRNA of the isolate used in study was taken as a template for cDNA synthesis using random decamer primers (Ambion, USA) and moloney murine leukemia virus–reverse transcriptase (Mo–MuLV–RT) enzyme (Sibzyme, Russia) as per the manufacturer’s protocol. The primer was allowed to anneal at 25°C for 10 minute followed by reverse transcription at 37°C for 60 minute and final inactivation at 90°C for 10 minute (Biorad i–Cycler).
The cDNA of isolate BT16/07 was subsequently subjected to PCR amplification using VP5 gene specific primer pairs of all the BTV serotype (along with primer P1 and P2). The amplification was carried out in 20 µl reaction mixture using phusion high–fidelity DNA polymerase (Finnzymes, Finland) in thermal cycler (Biorad iCycler, USA) as per manufacturer’s reaction protocol. The amplification programme consisted of initial denaturation for 2 min at 98°C, followed by 32 cycles for 10 seconds denaturation at 98°C and 30sec primer extension at 72°C. The annealing temperature was kept at 57°C for primer P1 and P2. Final primer extension was carried out at 72°C for 10 minute. The annealing temperature for group specific NS1 gene based PCR was fixed at 54°C for 30 seconds. The amplified PCR products were analysed using 1% agarose (Sigma, USA) gel electrophoresis.
Cloning of Partial VP5 Gene of BTV Isolate
The VP5 gene PCR product of the isolate in study was cloned in pJET1.2 vector using CloneJET™ PCR Cloning Kit (Fermentas, USA) as per the manufacturer’s protocol. The positive clones were selected based on colony touch PCR using VP5 gene specific primer. The overnight grown recombinant colonies containing VP5 gene of BT16/07 isolate were processed for plasmid isolation using QIAprep Spin Miniprep Kit (Qiagen, USA).
Nucleotide Sequencing and Data Analysis
The purified plasmid having VP5 gene of isolate in study was sequenced using plasmid specific primer as provided by manufacturer in Genetic Analyser ABI PRISMTM 3130 XL system in our laboratory. The terminal vector contamination was identified and removed using online tool VecScreen of GenBank (http://www. ncbi. nlm.nih. gov/ VecScreen /VecScreen .html). The VP5 gene sequence was subjected to BLAST search to GenBank data base for serotype confirmation using online tool BLASTn+ 2.2.28 (http://blast.ncbi.nlm.nih.gov/) (Zhang et al., 2005). The nucleotide sequence data was translated to deduced amino acid sequences according to its open reading frame. Bioedit v7.2.3 software (Hall, 1999) was used for calculation of percent identity of nucleotides as well as deduced amino acids of partial VP5 gene of BT16/07 isolate with previously reported isolates. The phylogenetic tree was constructed using neighbour joining method with 1000 bootstrap value in MEGA 5 software programme (Tamura et al., 2011). The in–silico restriction enzyme analysis (REA) of VP5 gene of BT16/07 isolate along with other isolates from different parts of world was carried out using Restriction Mapper version 3.0 software available online at http://www.restrictionmapper.org/.
RRESULTS
PCR for PCV2 Detection
The BT16/07 isolate was propagated in BHK–21 cell line. The virus isolate induced the characteristic CPE of BTV within 48 hours post infection, which was characterized by cellular rounding, foamy degeneration and death of infected cells resulting in detachment of cells from the surface of culture bottles. The dsRNA of the virus isolate was subjected to RNA–PAGE analysis. The migration pattern of BTV genomic dsRNA revealed the classical 10 segments with 3:3:3:1 migration pattern indicating that the sample was positive for BTV (Figure 1). The cDNA of the virus was subjected to the group–specific NS1 gene based PCR. The PCR yielded an expected amplicon size of 366 bp without any non–specific amplification indicating the sample as BTV (Figure 2).
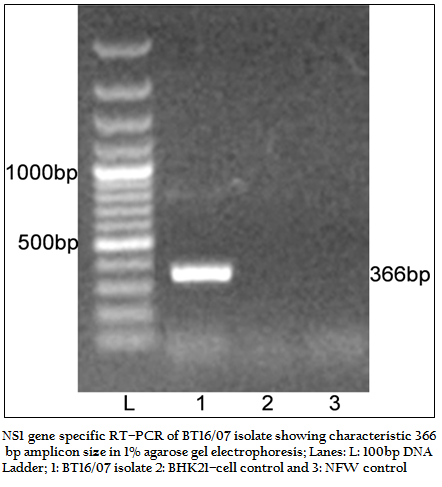
Figure 2:NS1 gene specific RT–PCR of BT16/07 isolate showing characteristic 366 bp amplicon size in 1% agarose gel electrophoresis; Lanes: L: 100bp DNA ladder; 1: BT16/07 isolate 2: BHK21–cell control and 3: NFW control.
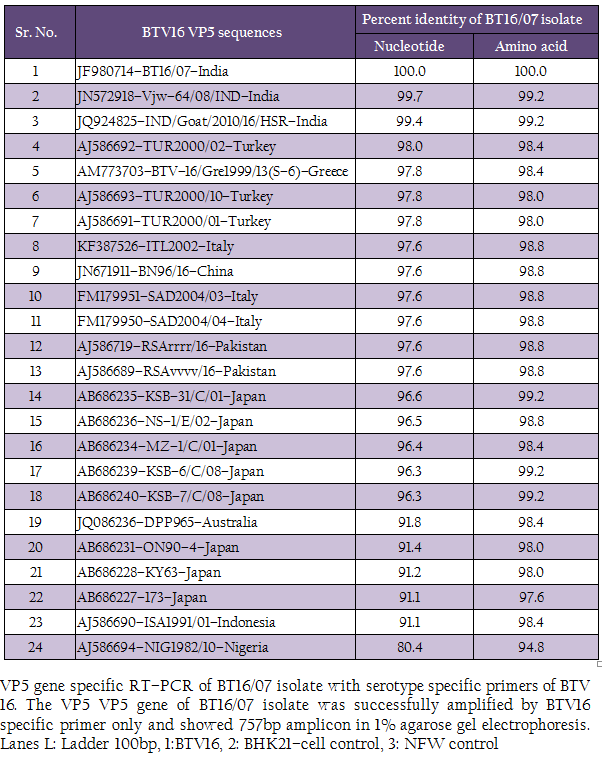
The cDNA of BT16/07 isolate was subjected to serotype specific PCR using VP5 gene specific primers of all the BTV serotypes. A single expected 757bp amplicon size was observed with BTV16 VP5 gene specific primer (P1 and P2) without any non–specific amplification (Figure 3). The remaining serotype specific primers did not show any amplification (data not shown). The cloned VP5 gene was subjected to sequencing. The nucleotide sequence obtained was deposited to GenBank database and an accession number JF980714 was assigned. The BLASTN+ 2.2.28 search revealed the maximum identity of the isolate in study with BTV16 from India (Accession number JN572918)
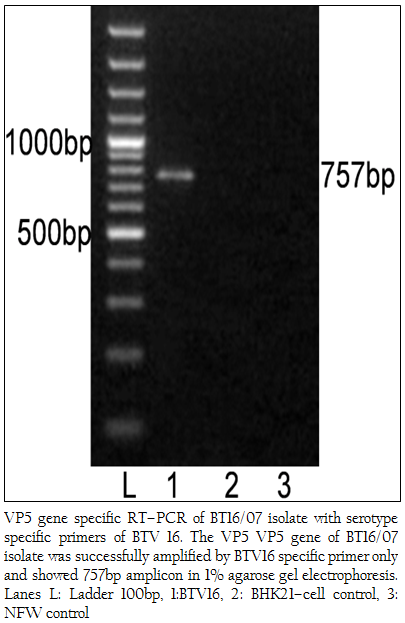
Figure 3: VP5 gene specific RT–PCR of BT16/07 isolate with serotype specific primers of BTV 16. The VP5 VP5 gene of BT16/07 isolate was successfully amplified by BTV16 specific primer only and showed 757bp amplicon in 1% agarose gel electrophoresis. Lanes L: Ladder 100bp, 1:BTV16, 2: BHK21–cell control, 3: NFW control
DISCUSSION
The BTV infection is often sub clinical, but can lead to severe disease with high mortality in susceptible ruminant animals. The presence of Culicoides vector, broad host range from domestic to wild animals, prevalence of multiple serotypes and non–availability of a suitable commercial vaccine are playing a major role in outbreaks of BT in India. The VP5 gene based phylogenetic relationship reference strains of all 24 serotypes have been reported earlier (Singh et al., 2004). However, VP5 gene based serotype confirmation and molecular analysis of BTV 16 of sheep origin from India has not been reported earlier. In present study the BTV isolate collected from Andhra Pradesh state (a state of high BTV prevalence) was serotyped and genetically characterised.
The BTV isolate BT16/07 (Accession number JF980714) produced CPE within 48 hours post infection in BHK 21 cell which is characterized by cell rounding, fusion, necrosis of infected cells and detachment from glass surface. The observed CPE indicated the presence of an infectious agent such as BTV, which required further confirmation. The characteristic CPE in BHK–21 cell culture, 3:3:3:1 migration pattern of viral genomic dsRNA in RNA–PAGE and specific amplicon of 366bp with group specific NS1 gene based PCR confirmed the virus isolate as BTV. The cDNA of the BT16/07 isolate (Accession number JF980714) was allowed for VP5 gene specific RT–PCR for serotype determination. A single expected 757bp PCR amplicon was observed with BTV16 VP5 gene specific primer (Figure 3). There was no any amplification observed with remaining serotype specific primers (data not shown). Thus, the isolate in study was serotyped as BTV16 based on VP5 gene specific RT–PCR.
The sequence data of cloned VP5 gene product revealed the maximum identity with BTV16 isolates on BLASTN+ 2.2.28 search, confirmed the virus isolate as BTV16. The sequence analyses revealed that Indian BT16/07 isolate share >99.0% nucleotide and amino acid identity with Indian isolates of BTV16 (GenBank accession number JQ924825 and JN572918). It showed 97.6% nucleotide and 98.8% amino acid identity with reference eastern BTV16 isolates from Pakistan (Maan et al., 2012b). The BT16/07 isolate showed 97.8 – 91.1% nucleotide and 99.2 – 97.6% amino acid identity with other eastern BTV16 isolates from Australia, Japan, Indonesia, China, Italy, Turkey, Greece isolates. However, it showed only 80.4% nucleotide and 94.8% amino acid identity with western BTV16 isolate from Nigeria. The nucleotide sequence analysis indicated the eastern origin BT16/07 isolate. The nucleotide sequence based phylogenetic analyses of BT16/07 isolate (Accession number JF980714) with other isolates of BTV16 showed major eastern and western clusters (Figure 4). However, BT16/07 isolate formed a much closer cluster with eastern isolates of BTV16 from India (Accession number JQ924825 and JN572918) Europe, Pakistan and China. The Australian isolate was distantly related to the major eastern cluster. The Japanese and Indonesian isolates formed a separate cluster and distantly related to Indian isolates. The Nigerian isolate formed a separate western cluster of BTV16. Thus, the phylogenetic analysis also indicated the eastern origin of BT16/07 isolate.
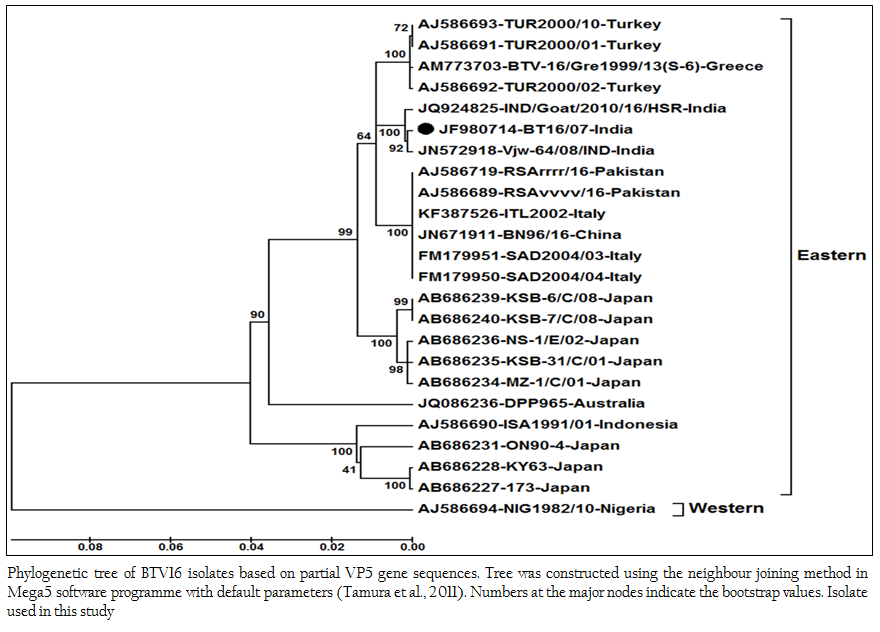
Figure 4: Phylogenetic tree of BTV16 isolates based on partial VP5 gene sequences. Tree was constructed using the neighbour joining method in Mega5 software programme with default parameters (Tamura et al., 2011). Numbers at the major nodes indicate the bootstrap values. Isolate used in this study.
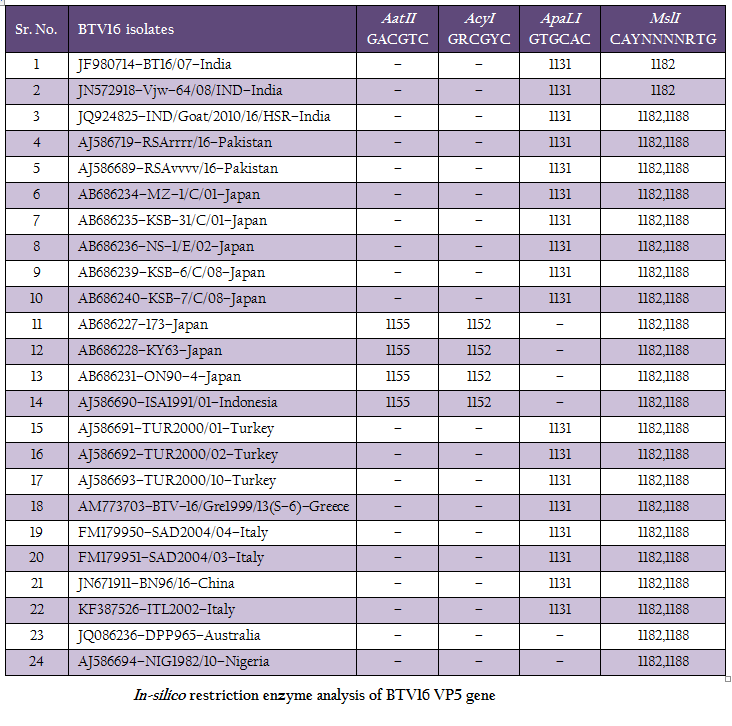
The RT–PCR in combination with REA has been found very useful tool in molecular characterization of BTV isolates directly from clinical samples. The in–silico REA of VP5 gene of Indian BT16/07 isolate (Accession number JF980714) and other isolates of BTV16 available in GenBank was done using web based tool restriction mapper version 3.0 (Table 2). The in– silico REA with ApaLI restriction enzyme revealed the single restriction site at nt 1131 in all the Indian (including BT16/07 isolate), European (Turkey, Greece and Italy), Pakistan, Japan and China isolates. The same ApaLI restriction enzyme did not show any restriction site in BTV16 isolates from Indonesia, Australia, some isolates from Japan (Accession number AB686227, AB686228 and AB686231). The western BTV16 isolate from Nigeria also did not show any restriction site with ApaLI restriction enzyme. Similarly, in–silico REA with AcyI and AatII enzymes revealed a single restriction site at nt1152 and nt 1155 respectively in BTV16 isolates from Indonesia and Japan (Accession number AB686227, AB686228 and AB686231). However, the AcyI and AatII enzymes did not show any restriction sites in any other BTV16 isolates includingBT16/07 isolate. The in–silico REA using ApaLI, AcyI and AatII enzymes revealed the closeness of BT16/07 isolate to eastern BTV16 isolates from different part of globe. The in– silico REA with MslI enzyme revealed a single restriction site at nt 1182 in Indian isolates BT16/07 (Accession number JF980714) andVjw–64/08/IND (Accession number JN572918). However, the same MslI enzyme revealed the two restriction sites at nt1182 and1188 in all the Asian, European, Australian and Nigerian isolates of BTV16. Therefore, REA using MslI enzyme can be used for differentiation of Indian BT16/07 and Vjw–64/08/IND (Accession number JN572918) isolate to the global BTV16 isolates.
The BTV16 has been reported earlier from Andhra Pradesh (Shafiq et al., 2013) and Tamil Nadu state (Minakshi et al., 2012) of south India. The BTV isolate in study (BT16/07) has been reported from sheep in Andhra Pradesh state, the adjoining state of Tamil Nadu. The high nucleotides as well as amino acid identity (>99%) between BT16/07 (Accession number JF980714)and BTV16 isolate from Andhra Pradesh (Accession number JN572918) and Tamil Nadu (Accession number JQ924825) isolates revealed close origin of these viruses. Since south India particularly Andhra Pradesh and Tamil Nadu is endemic for a major species of vector i.e. Culicoides oxystoma. Therefore, the same BTV16 serotype might be migrated from Tamil Nadu to Andhra Pradesh or vice versa either through vector or migrating sheep population.
The RT–PCR, nucleotide sequencing, phylogenetic analysis and in–silico restriction analysis of VP5 gene could be helpful in serotyping and molecular characterization of BTV. The BTV isolate used in study was confirmed as BTV 16 based on VP5 gene RT–PCR and nucleotide sequencing. The VP5 gene specific primer tested can be used for identifying the BTV16 serotype prevalent in endemic region and suitable vaccine candidate can be identified for controlling the disease. These findings also suggested that sequence based REA tools could be effectively used for differentiation of Indian isolates from rest of the global BTV16 isolates.
CONCLUSIONS
A new BT Virus (BT16/07) was isolated from Andhra Pradesh state of India. The new isolate was confirmed as BTV serotype 16 based on VP5 gene specific PCR amplification and nucleotide sequencing. The phylogenetic analysis and in–silico restriction enzyme analysis confirmed that BT16/07 isolate belong to eastern topotype. BT16/07 isolate was found very closer (>99% nucleotide and amino acid identity) to other Indian BTV16 isolates. The VP5 gene specific primer tested in this study can be used for molecular characterization of BTV16 serotype prevalent in endemic region.
CONFLICT OF INTEREST
There is no conflict of interest regarding study.
ACKNOWLEDGEMENTS
This study was funded by Indian Council of Agricultural Research, New Delhi under ‘All India network programme on Bluetongue’. The authors are thankful to Department of Animal Biotechnology, LLR University of Veterinary and Animal Sciences, Hisar for providing infrastructural facility.
REFERENCES
Belhouchet M, MohdJaafar F, Firth AE, Grimes JM, Mertens PPC (2011). Detection of a fourth Orbivirus non–structural protein. PloS. ONE 6: e25697.
http://dx.doi.org/10.1371/journal.pone.0025697
PMid:22022432 PMCid:PMC3192121
Darpel KE, Batten CA, Veronesi E, Shaw AE, Anthony S, Bankowska BK, Kgosana L, bin–Tarif A, Carpenter S, Müller–Doblies U, Takamatsu H, Mellor PS, Mertens PPC, Oura CAL (2007). Clinical signs and pathology shown by British sheep and cattle infected with bluetongue virus serotype 8 derived from the 2006 outbreak in northern Europe. Vet. Rec. 161: 253 – 261.
http://dx.doi.org/10.1136/vr.161.8.253
PMid:17720961
Gollapalli SR, Mallavarapu S, Uma M, Rao PP, Susmitha B, Prasad PU, Chaitanya P, Prasad G, Hegde NR, Reddy YN (2012). Sequences of genes encoding type–specific and group–specific antigens of an Indian isolate of bluetongue virus serotype 10 (BTV–10) and implications for their origin. Transbound. Emerg. Dis. 59(2): 165 – 172.
http://dx.doi.org/10.1111/j.1865-1682.2011.01266.x
PMid:22032621
Hall TA (1999). BioEdit: a user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41: 95 – 98.
Hofmann MA, Renzullo S, Mader M, Chaignat V, Worwa G, ThuerB (2008). Genetic characterization of Toggenburgorbi virus a new bluetongue virus from goats Switzerland. Emerg. Infect. Dis. 121: 855 – 1861.
Howerth EW, Greene CE, Prestwood AK (1988). Experimentally induced bluetongue virus infection in white tailed deer: coagulation, clinical pathologic and gross pathologic changes. Am. J. Vet. Res. 49: 1906 – 1913.
PMid:2854709
Kovi RC, Dahiya S, Minakshi, Prasad G (2005). Evaluation of new primers targeting serogroup specific genes for detection of bluetongue viruses by RT–PCR. Indian J. Microbiol. 45: 103 – 119.
Laemmli UK (1970). Cleavage of structural proteins during the assembly of head of bacteriophage T4. Nature 227: 680 – 685.
http://dx.doi.org/10.1038/227680a0
PMid:5432063
Maan S, Maan NS, Nomikou K, Eva V, Bankowska KB, Manjunatha BN, Houssam A, Mertens PPC (2011). Complete Genome Characterisation of a Novel 26th Bluetongue Virus Serotype from Kuwait. PlosOne 6: 1 – 11.
http://dx.doi.org/10.1371/journal.pone.0026147
PMid:22031822 PMCid:PMC3198726
Maan S, Maan NS, Pullinger G, Nomikou K, Morecroft E, Guimera M, Belaganahalli MN, Mertens PP (2012a). The genome sequence of bluetongue virus type 10 from India: evidence for circulation of a western topotype vaccine strain. J Virol. 86(10): 5971 – 5972.
http://dx.doi.org/10.1128/JVI.00536-12
http://dx.doi.org/10.1128/JVI.00596-12
PMid:22532535 PMCid:PMC3347277
Maan S, Maan NS, Singh KP, Belaganahalli MN, Guimera M, Pullinger G, Nomikou K, Mertens PPC (2012b). Complete genome sequence analysis of a reference strain of Bluetongue virus serotype 16. J. Virol. 86(18):10255 – 10256. DOI: 10.1128/JVI.01672–12.
http://dx.doi.org/10.1128/JVI.01672-12
Maclachlan NJ, Drew CP, Darpel KE, Worwa G (2009). The Pathology and Pathogenesis of Bluetongue. J. Comp. Path. 141: 1 – 16.
http://dx.doi.org/10.1016/B978-012369368-6.50017-4
Mellor PS (1990). The replication of bluetongue virus in Culicoides vectors. Curr. Top. Microbiol. Immunol.162: 143 – 161.
http://dx.doi.org/10.1007/978-3-642-75247-6_6
Mertens PPC, Maan S, Samuel A, Attoui H (2004). Orbivirus Reoviridae. In: Virus Taxonomy, VIIth Report of the ICTV, edited by C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, L.A. Ball (London: Elsevier/Academic Press), 466 – 483.
Mertens PPC, Pedley S, Cowley J, Burroughs JN, Corteyn AH, Jeggo MH, Jennings AM, Gorman BM (1989). Analysis of the roles of bluetongue virus outer capsid proteins VP2 and VP5 in determination of virus serotype. Virol. 170: 561 – 565.
http://dx.doi.org/10.1016/0042-6822(89)90447-9
Minakshi P, Singh R, Ranjan K, Kumar P, Joshi CG, Reddy YKM, Prasad G (2012). Complete Genome Sequence of Bluetongue Virus Serotype 16 of Goat Origin from India. J. Virol. 86: 8337 – 8338.
http://dx.doi.org/10.1128/JVI.01128-12
PMid:22787269 PMCid:PMC3421637
Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MP, Summers MD (1995). Virus taxonomy, classification and nomenclature of viruses. In Sixth Report of the International Committee on Taxonomy of Viruses. Arch. Virol. 10: 412 – 414.
Prasad G, Minakshi, Malik Y (2007). 'Bluetongue' (Eds: G. Prasad, Minakshi and Yashpal Malik). A book published by Indian Council of Agricultural Research, New Delhi.
Ratinier M, Caporale M, Golder M, Franzoni G, Allan K, Nunes SF, Armezzani A, Bayoumy A, Rixon F, Shaw A, Palmarini M (2011). Identification and Characterization of a Novel Non–Structural Protein of Bluetongue Virus. PLoSPathog 7(12): e1002477. doi:10.1371/journal.ppat.1002477.
http://dx.doi.org/10.1371/journal.ppat.1002477
Singh KP, Maan S, Samuel AR, Rao S, Meyer AJ, Mertens PPC (2004). Phylogenetic analysis of bluetongue virus genome segment 6 (encoding VP5) from different serotypes. Vet. Ital. 40: 479 – 483.
PMid:20422573
Susmitha B, Sudheer D, Rao PP, Uma M, Prasad G, Minakshi P, Hegde NR, Reddy YN (2012). Evidence of bluetongue virus serotype 21 (BTV–21) divergence. Virus Genes. 44(3): 466 – 469.
http://dx.doi.org/10.1007/s11262-012-0724-y
PMid:22350945
Svensson L, Uhnoo I, Grandien M, Wadeli G (1986). Molecular epidemiology of rotavirus infections in Upsala. Sweden. 1981; disappearance of a predominant electropherotype. J. Med. Virol. 18: 101 – 111.
http://dx.doi.org/10.1002/jmv.1890180202
PMid:3005484
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28: 2731 – 2739.
http://dx.doi.org/10.1093/molbev/msr121
PMid:21546353 PMCid:PMC3203626
Zhang Z, Schwartz S, Wagner L, Miller W (2000). A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203 – 214.
http://dx.doi.org/10.1089/10665270050081478
PMid:10890397