Advances in Animal and Veterinary Sciences
Research Article
Virulence Potential, Antimicrobial Susceptibility and Phylogenetic Analysis of Corynebacterium pseudotuberculosis Isolated from Caseous Lymphadenitis in Sheep and Goats in Duhok City, Iraq
Rezheen F. Abdulrahman*
Pathology and Microbiology Department, Collage of Veterinary Medicine, University of Duhok, Duhok, Kurdistan Region, Iraq.
Abstract | Caseous lymphadenitis is a bacterial disease caused by Corynebacterium pseudotuberculosis affecting sheep and goats worldwide. It has been previously shown that the CLA is highly prevalent in sheep and goats in Duhok city. Therefore, this study is aimed to study virulence genes, the susceptibility pattern toward antimicrobial agents and to analyse the phylogenetic relationship of C. pseudotuberculosis based on the partial rpoB gene sequences. The results showed that the virulence genes including pld, fagA, B, C and D were identified 100% in all 22 isolates used in this study. Antibiotics sensitivity test showed varying degrees of susceptibility patterns against cefotaxime, clarithromycin, gentamicin, rifampin, amoxicillin and trimethoprim/sulphamethoxazole. The isolates were found to be sensitive 100% to chloramphenicol, doxycycline, norfloxacin, ciprofloxacin, clindamycin, erythromycin, ceftriaxone, levofloxacin. The overall degree of resistance was very low, with the exception of amoxicillin which was 81.8%. Nucleotide sequence analysis of the 22 rpoB sequences detected the overall similarity of 97, 98, 99 and 100% with biovar ovis of the reference sequences. Phylogenetic tree of partial sequences of the rpoB gene showed that the biovar ovis isolates from both sheep and goat are grouped together in the same lineage and the nucleotide variation between the sequences does not change the protein expression. In conclusion, for the first time in Duhok and in Iraq, phylogenetic tree clearly confirmed the C. pseudotuberculosis subspecies differentiation based on rpoB sequences from sheep and goats with CLA.
Keywords | C. pseudotuberculosis, Virulence; antibiotic, Phylogenetic analysis, Sheep and goat, Duhok
Received | March 12, 2021; Accepted | March 17, 2021; Published | May 25, 2021
*Correspondence | Rezheen F. Abdulrahman, Pathology and Microbiology Department, Collage of Veterinary Medicine, University of Duhok, Duhok, Kurdistan Region, Iraq; Email: [email protected]
Citation | Abdulrahman RF (2021). Virulence potential, antimicrobial susceptibility and phylogenetic analysis of corynebacterium pseudotuberculosis isolated from caseous lymphadenitis in sheep and goats in duhok city, iraq. Adv. Anim. Vet. Sci. 9(6): 919-925.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.6.919.925
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Abdulrahman. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
C. pseudotuberculosis is a Gram-positive, pleomorphic, non-motile, non-capsulated, non-spore forming, facultative intracellular microorganism causing caseous lymphadenitis (CLA) in sheep and goats (Baird and Fontaine, 2007; Guimarães et al., 2011; Umer et al., 2017; Parin et al., 2018; Guerrero et al., 2018). C. pseudotuberculosis is classified into two biovars according to nitrate reduction activity including biovar ovis and biovar equi (Connor et al., 2000; Connor et al., 2007; Oliveira et al., 2016; Guerrero et al., 2018). The biovar ovis is a cause of CLA in sheep and goats, while biovar equi causes disease in horses, cattle and buffalo (Dorella et al., 2006; Ruiz et al., 2011; Oliveira et al., 2016). The disease is characterized by abscesses formation in lymph nodes and visceral organs (Çetinkaya et al., 2002; Markey et al., 2013; Umer et al., 2017). C. pseudotuberculosis possesses variety of virulence factors that are involved in the pathogenesis of the disease. However, the mechanism of infection is still poorly understood (Dorella et al., 2006). Recently virulence genes related to the pathogenicity of C. pseudotuberculosis have been identified using whole genome analysis allowing understanding pathogenicity of this bacterium (Ruiz et al., 2011). Ruiz et al. (2011) reported the presence of seven putative pathogenicity islands containing several classical virulence factors, including genes encoding adhesion factors, fimbrial subunits, iron uptake and secreted toxins and these genes probably acquired via horizontal gene transfer. The main virulence factor of C. pseudotuberculosis is phospholipase D (plD) which is an exotoxin that increases vascular permeability and promotes the hydrolysis of cell membrane of the host that facilitates the dissemination of the pathogen from the primary site to secondary sites. Thus, allows bacterial survival in the cells, the invasion of the body and transport to regional lymph nodes by phagocytic cells (Dorella et al., 2006; Baird and Fontaine, 2007). Other virulence genes including fagA, B, C and D are involved in the pathogenesis of C. pseudotuberculosis and they encode putative iron uptake system allowing intracellular survival of this bacterium (Dorella et al., 2006). It has been previously shown that the CLA is highly prevalent in sheep and goats in Duhok city (Abdulrahman et al., 2020; Issa et al., 2021). However, to our knowledge yet no study has been shown basic information regarding virulence determinant and controlling this pathogen in Duhok. Moreover, no studies have shown the phylogenetic analysis of C. pseudotuberculosis isolates based on rpoB from CLA from sheep and goats in Iraq. Therefore, the present study was undertaken to identify the presence of virulence genes, to study the susceptibility pattern to antimicrobial agents and to analyse the phylogenetic relationship of C. pseudotuberculosis isolates from slaughtered sheep and goats suffering from CLA in Duhok city.
MATERIALS AND METHODS
Sample Collection and Detection of C. Pseudotuberculosis
From1090 of inspected carcasses of sheep and goats slaughtered at Duhok abattoir, 22 were infected with CLA (Abdulrahman et al., 2020; Issa et al., 2021). All suspected cases of CLA were found to be positive with C. pseudotuberculosis. Isolation of C. pseudotuberculosis was based on traditional bacteriological methods using colony morphologies, Gram staining, biochemical reactions. Suspected isolates were confirmed by amplification of the specific C. pseudotuberculosis target genes as carried out in our previous studies (Abdulrahman et al., 2020; Issa et al., 2021). Isolation and molecular characterization were carried out at Duhok research Centre, College of Veterinary Medicine, University of Duhok.
Detection Of Virulence Genes
C. pseudotuberculosis isolates were plated out onto 5% sheep blood agar from glycerol stocks and incubated at 37°C for 24 h. DNA was extracted as described by (de Sá et al., 2013). The following virulence genes were selected: phospholipase D (plD), intergral membrane protein (fagA), iron enterobactin transporter (fagB), ATP binding cytoplasmic membrane protein (fagC) and iron siderophore binding protein (fagD). The final volume of 10 μL of PCR reaction was used. Each reaction contains 5.5 μL ready-to-use master mixes (Ruby Taq Master®, Jena Bioscience, Thuringia, Germany), 2.5 μL genomic DNA, 1 μL of each primer (10 pmol). Primers used for the detection of virulence genes are shown in Table 1 and they were synthesized by Macrogen Company (South Korea). The amplification of 40 cycles was performed in a Gene Amp PCR System 9700 Thermo Cycler (Applied Biosystems) according to (de Sá et al., 2013): initial denaturation of 94 °C for 30 s, denaturation at 94 °C for 40 s, annealing for 40 s at 61°C, 58°C, 55°C, 60°C and 61°C for the plD, fagA, fagB, fagC and fagD, respectively, extension at 72 °C for 40 s and a final extension to 72 °C for 10 min. The estimated size of amplified products for each gene (Table 1) was detected in 1% agarose gels stained with Prime Safe Dye (GeNet Bio, Korea) and viewed under UV light.
Antimicrobial Susceptibility Test of C. Pseudotuberculosis
The sensitivity patterns of C. pseudotuberculosis isolates against antimicrobial agents was tested using the Kirby-Bauer-disk diffusion method according to Clinical Laboratory and Standards Institute (CLSI) (CLSI, 2015). Yet, no specific guideline has been published form CLSI for antimicrobial susceptibility test of C. pseudotuberculosis (Dorella et al., 2006). Therefore, the following antibiotics with concentration (µg/disc) were selected: chloramphenicol (30), cefotaxime (30), doxycycline (30), clarithromycin (15), gentamicin (10), norfloxacin (10), ciprofloxacin (5), clindamycin (2), rifampin (5), amoxicillin (10), erythromycin (15), ceftriaxone (30), levofloxacin (5) and trimethoprim/sulphamethoxazole (25) (Bioanalyse). According to manufacturer’s instructions, the susceptibility of the isolates to the antibiotics was measured according to the size of the inhibition zone.
Ropb Gene Amplification and Sequencing
The primer pair C2700-F (5´-CGTATGAACATCGGCCAGGT-3´) and C3130-R (5´-TCCATTTCGCCGAAGCGCTG-3´) was used to amplify partial sequence of the rpoB individually according to (Khamis et al., 2004). Primers were synthesised by Macrogen (Seoul, South Korea). A total reaction volume of 50 µl was used. Each reaction contains the following reagents: 25 µl of ready-to-use master mixes (Ruby Taq Master®, Jena Bioscience, Thuringia, Germany), 2 µl of each forward and reverse primer (10 pmol µl-1), 5 µl of template DNA and 16 µl dH2O. Amplification was performed in a Gene Amp PCR System 9700 Thermo Cycler (Applied Biosystems) using 35 cycles of the following amplification parameters: initial denaturation at 94°C for 3 min, denaturation
Table 1: Details of oligonucleotides primers for detection of C. pseudotuberculosis virulence gene
| Virulence gene | Primer name | Sequence (5'—3΄) | Product size (bp) | Ref |
|
plD
|
Pld-F | ATG AGG GAG AAA GTT GTT TTA | 924 | |
| Pld-R | TCA CCA CGG GTT ATC CGC | |||
|
fagA |
FagA_F | AGC AAG ACC AAG AGA CAT GC | 245 | |
| FagA_R | AGT CTC AGC CCA ACG TAC AG | |||
|
fagB |
FagB_F | GTG AGA AGA ACC CCG GTA TAA G | 291 | |
| FagB_R | TAC CGC ACT TAT TCT GAC ACT G | |||
|
fagC |
FagC_F | GTT TGG CTA TCT CCT TGG TAT G | 173 | |
| FagC_R | CGA CCT TAG TGT TGA CAT ACC C | |||
|
fagD |
FagD_F | GAG ACT ATC GAC CAG GCA GA | 226 | |
| FagD_R |
ACT TCT TGG GGA GCA GTT CT |
Table 2: Susceptibility profile of C. pseudotuberculosis isolates from sheep and goat with CLA (no: 22)
|
Antibiotics
|
Concentrations (µg/disc)
|
R | IM | S | |||
| No. of isolates | Rate (%) | No. of isolates | Rate (%) | No. of isolates | Rate (%) | ||
| Chloramphenicol | 30 | 0 | 0% | 0 | 0% | 22 | 100% |
| Cefotaxime (CTX) | 30 | 0 | 0% | 6 | 27.3% | 16 | 72.7% |
| Doxycycline (DO) | 30 | 0 | 0% | 0 | 0% | 22 | 100% |
| Clarithromycin (CLR) | 15 | 0 | 0% | 2 | 9.1% | 20 | 90.9% |
| Gentamicin (CN) | 10 | 1 | 4.5% | 3 | 13.7% | 18 | 81.8% |
| Norfloxacin (NOR) | 10 | 0 | 0% | 0 | 0% | 22 | 100% |
| Ciprofloxacin (CIP) | 5 | 0 | 0% | 0 | 0% | 22 | 100% |
| Clindamycin (DA) | 2 | 0 | 0% | 0 | 0% | 22 | 100% |
| Rifampin (RA) | 5 | 2 | 9.1% | 2 | 9.1% | 18 |
81.8.% |
| Amoxicillin (AX) | 10 | 18 | 81.8% | 2 | 9.1% | 2 | 9.1% |
| Erythromycin € | 15 | 0 | 0% | 0 | 0% | 22 |
100% |
| Ceftriaxone (CRO) | 30 | 0 | 0% | 0 | 0% | 22 | 100% |
| Levofloxacin (Lev) | 5 | 0 | 0% | 0 | 0% | 22 | 100% |
| Trimethoprim/sulphamethoxazole (SXT) | 25 | 3 | 13.6% | 4 | 18.2% | 15 |
68.2% |
R: resistance; IM: intermediate; S: Sensitive
at 94°C for 1 min, annealing at 56°C for 1 min, extension at 72°C for 2 min and a final extension step at 72°C for 7 min. The purity and concentration of amplified products were examined by 1% (w/v) agarose gel electrophoresis and NanoDrop 2000C spectrophotometer (Thermo Scientific/ UK). Finally, 40 µl of amplified was sequenced using Sanger sequencing technique by Macrogen, inc Sequencing Service (Seoul, South Korea).
Sequence Analysis and Phylogenetic Tree
The partial sequences of the rpoB were received as .ab1 chromatogram files. The sequence files were viewed, trimmed and analysed using the BioEdit sequence alignment editor version7 (Hall, 1999). Sequencing errors were also checked and corrected in BioEdit. Multiple sequence alignment with ClustalW and phylogenetic analyses were conducted using Molecular Evolutionary Genetics Analysis (MEGA7) software (Kumar et al., 2016). MEGA7 was used to generate Neighbour-Joining phylogenetic trees using the Jukes-Canter correction model and Bootstrap (1000 replications) analysis. Basic Local Alignment Search Tool (BLAST) analysis was used for GenBank database searching and also to determine the similarity of the sequenced rpoB with references sequences in GenBank database.
RESULTS AND DISCUSSION
Virulence Genes Among C. Pseudotuberculosis
From 1090 of inspected carcasses of sheep and goats slaughtered at Duhok abattoir, twenty two were infected with CLA (Abdulrahman et al., 2020; Issa et al., 2021). The twenty two CLA cases were found to be positive with C. pseudotuberculosis as mentioned in our previous studies based on phenotypic and molecular characterizations (Abdulrahman et al., 2020; Issa et al., 2021). The twenty two isolates were examined for the presence of the virulence genes including pld, fagA, fagB, fagC and fagD genes. The results showed that all the virulence genes were detected in the 22 C. pseudotuberculosis isolates as shown in Fig. 1.
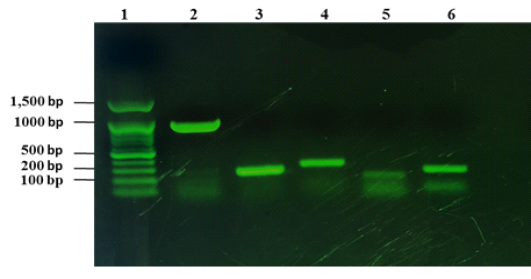
Figure 1: Detection of virulence genes of C. pseudotuberculosis isolates from sheep and goats with CLA. Lane 1: 100 bp DNA marker, lane 2: amplification of 924 bp of pld, lane 3: amplification of fagA (245 bp), lane 4: amplification of 291 bp of fagB, lane 5: amplification of fagC (173 bp), lane 6: amplified product of fagD gene (226 bp).
Antimicrobial Profile of C. Pseudotuberculosis Isolates
Fourteen antibiotics were tested against C. pseudotuberculosis isolates and the antibiotic susceptibility patterns are shown in Table 2. As shown in Table 2, variation was observed in susceptibility with cefotaxime, clarithromycin, gentamicin, rifampin, amoxicillin and trimethoprim/sulphamethoxazole. All isolates were found to be sensitive 100% to chloramphenicol, doxycycline, norfloxacin, ciprofloxacin, clindamycin, erythromycin, ceftriaxone, levofloxacin. However, cefotaxime, clarithromycin, gentamicin, rifampin, and trimethoprim/sulphamethoxazole showed variation in susceptibility rate at 72.7%, 90.9%, 81.8%, 81.8% and 68.2%, respectively. Low degree of susceptibility was also found with cefotaxime, clarithromycin, gentamicin, rifampin, amoxicillin and trimethoprim/sulphamethoxazole at rate of 27.3%, 9.1%, 13.7%, 9.1%, 9.1% and 18.2, respectively. The overall degree of resistance of the isolated C. pseudotuberculosis was very low, with the exception of amoxicillin (81.8%) as listed in Table 2.
Phylogenetic Analysis of C. Pseudotuberculosis Isolates
Twenty two partial sequence of rpoB gene were obtained as mentioned above in ropB gene amplification and sequencing. Twenty two Partial sequences of rpoB sequenced in this study have been deposited in the GenBank database under accessions number: MT974521 to MT974542). Nucleotide sequence comparison of partial rpoB sequences using BLAST analysis against globally published C. pseudotuberculosis showed that sequences similarity ranged from 97, 98, 99 and 100% especially with C. pseudotuberculosis biovar ovis from sheep and goats with CLA. Therefore, to generate phylogenetic tree in addition to partial rpoB sequences (C. pseudotuberculosis RA1-RA22) obtained in this study, biovar ovis (GenBank accessions number: CP001809.3, CP015100.2, CP001829.2 and CP047603.1) were included. To differentiate between biovar ovis and equi, reference sequences of biotypes equi (GenBank accessions number: CP003540.3 and CP003652.3) were also added. To create out group in phylogenetic tree, reference sequence of C. ulcerans (GenBank AY492271.1) were included to perform analysis as shown in Fig. 2. The rpoB phylogenetic tree showed that 22 rpoB sequences were grouped with C. pseudotuberculosis biovar ovis. Phylogenetic analyses of rpoB sequences can clearly distinguish biovar ovis from biovar equi as shown Fig. 2. Comparative sequence analysis of the 22 rpoB sequences (398bp and 132 amino acids) showed that there were total 9 polymorphic nucleotide sites and 5 variable amino acid sites. Pairwise differences ranged from 1 to 8 nucleotide sites and from 1 to 4 amino acid positions. Pairwise differences do not modify the expression of protein and it clearly shown that rpoB gene from ovine and caprine isolates were grouped in the same lineage (Fig. 2).
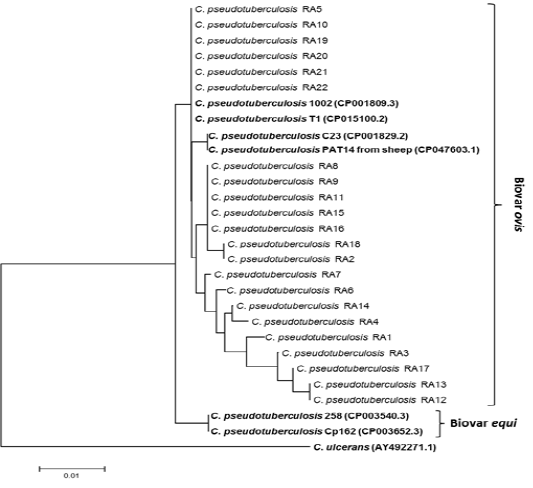
Figure 2: Phylogenetic tree of C. pseudotuberculosis isolates based on the rpoB gene sequence. The tree was conducted with MEGA7 using Neighbor-joining method and Bootstrap value of 1000 repetitions. Twenty two rpoB genes of the C. pseudotuberculosis isolates obtained in this study designated as C. pseudotuberculosis RA1-RA22 as shown in tree and they were deposited in the GenBank database under accessions number: MT974521 to MT974542).
DISCUSSION
C. pseudotuberculosis is a facultative intracellular pathogen causing CLA mainly in sheep and goats and it causes economic loses worldwide. The results of our previous studies showed that the prevalence of CLA in sheep and goats is high is Duhok city (Abdulrahman et al., 2020; Issa et al., 2021). Similarly, CLA is highly prevalent in small ruminants globally. Consequently, this study reported for the first time the investigation of virulence genes, antibiotic susceptibility and phylogenetic analysis of 22 C. pseudotuberculosis isolates from slaughtered sheep and goats suffering from CLA for better understanding the epidemiology of C. pseudotuberculosis in Duhok ciry, Iraq.
In this study, five important virulence genes including pld, fagA, B, C and D were selected to confirm their presence in C. pseudotuberculosis isolates. The pld encodes the phospholipase D-PLD exotoxin enzyme that causes increasing vascular permeability and facilitates the spreading of the pathogen within the host (Dorella et al., 2006; Baird and Fontaine, 2007). The results showed that pld gene was found in all isolated C. pseudotuberculosis characterized in this study. The results were in agreement with other studies (Çetinkaya et al., 2002; Pacheco et al., 2007; de Sá et al., 2013; Guerrero et al., 2018; Li et al., 2018). The mechanism of pld gene expression still understood. However, comparative proteomic analyses of the two C. pseudotuberculosis strains from sheep and goat with CLA revealed the expression of this protein only in virulent isolate (Pacheco et al., 2011). A study by McNamara et al. (1994) confirmed that pld gene is a virulence determinant of C. pseudotuberculosis that increases the maintenance and spread of the bacteria within the host because and they showed that the mutation of pld gene decreased the virulence of C. pseudotuberculosis and reduced ability to cause CLA.
The fagA, B, C and D encode putative iron uptake system allowing intracellular survival of C. pseudotuberculosis and they are organized in an operon involved in iron uptake which is located downstream from the pld gene and are expressed rarely in vitro (Billington et al., 2002). However, they increase the virulence of C. pseudotuberculosis when expressed in vivo indicating that the expression of these genes may be required host factors (Billington et al., 2002). The results of this study confirmed the presence fagA, B, C and D were 100% in all isolates. Similarly, a study by Li et al. (2018) found that fagA, B, C and D were found 100% in all 40 C. pseudotuberculosis isolates from goats with CLA.
Likewise, fag A, fagC, and fagD virulence genes were identified in the 100% and the fagB was only identified in 56 (98.2%) of the C. pseudotuberculosis isolates (57/57) from sheep and goats with CLA superficial abscesses (Guerrero et al., 2018). In a study by de Sá et al. (2013) of 168 C. pseudotuberculosis isolates from sheep and goats with CLA (140 isolates from superficial lesions, and 23 from visceral lesions) were examined, fagA, and fagB were identified in the 100% and fagC detected in the 99.4% (167/168) of examined isolates. However, regarding fagD the results were different and it was identified only in the 95.2% (160/168) and interestingly, these isolates were obtained from CLA superficial abscesses (de Sá et al., 2013). The C. pseudotuberculosis isolates analysed in this study were isolates from mediastinal CLA (visceral form) and similarly the virulence genes were identified 100% in the isolates from animals with visceral form of CLA (de Sá et al., 2013). The differences of virulence factors between visceral and superficial CLA suggests differences in the invasion prospective of the C. pseudotuberculosis strains. The presence of pld, fagA, B, C and D in the 100% of the C. pseudotuberculosis isolates indicating high virulence potential and those genes could be used as candidates for vaccine production (Billington et al., 2002; Pacheco et al., 2011).
Although the disease occurs worldwide causing major economic losses in sheep and goat industry areas, no effective treatment exists and yet no specific guideline has been published form CLSI for antimicrobial susceptibility test of C. pseudotuberculosis. Therefore, in the current study the efficiency of 14 antibiotics were tested against C. pseudotuberculosis isolates. All isolates were found to be sensitive 100% to chloramphenicol, doxycycline, norfloxacin, ciprofloxacin, clindamycin, erythromycin, ceftriaxone, levofloxacin. However, cefotaxime, clarithromycin, gentamicin, rifampin, and trimethoprim/sulphamethoxazole showed variation in susceptibility. The overall resistance degree was very low, with the exception of amoxicillin (Table 2). Sensitivity of the C. pseudotuberculosis isolates against different antimicrobial has been reported previously in many studies (Connor et al., 2007; Abebe and Sisay Tessema, 2015; Algammal, 2016; Li et al., 2018).
The 16S rRNA gene sequence was used most commonly for identification of bacteria and phylogeny as molecular marker. However, high polymorphism area in the rpoB gene was identified and used for the identification of C. pseudotuberculosis from clinical samples (Khamis et al., 2004). It was found that the rpoB is a more suitable sequence to differentiate C. pseudotuberculosis subspecies than 16S rRNA because of higher degree of nucleotide polymorphism (Khamis et al., 2004). Therefore, the analysis of polymorphisms in the partial rpoB sequence was used as a diagnostic tool that differentiates C. pseudotuberculosis strains at subspecies level (Retamal et al., 2011). In the present study sequencing of the partial rpoB showed that the 22 isolates confirmed as C. pseudotuberculosis based on BLAST anlysis and they were 97, 98, 99 and 100% similar to the C. pseudotuberculosis biovar ovis. Phylogenetic analysis of the 22 partial rpoB (Fig. 2) grouped ovine and caprine in the same clade and they grouped with biovar ovis from reference sequences. The results of the current study revealed that the rpoB differentiates biovar ovis from biovar equi because they are grouped into two different clades. Similarly a study by Guerrero et al. (2018) confirmed that the phylogenetic analysis based on the rpoB gene confirmed the subspecies differentiation and allowed the identification of point mutation in caprine isolates without modifying the expression of protein. In contrast, high polymorphism of the rpoB gene allowed caprine and ovine isolates into different lineages and the differences in the grouping could be due to the limited number of isolates and small geographical area (Retamal et al., 2011). Another housekeeping gene was also used as a candidate for differentiation of subspecies ovis form equi as shown by Li et al. (2018).
CONCLUSION
The results of the present study revealed that virulence genes including pld, fagA, B, C and D were identified in the all C. pseudotuberculosis isolated from sheep and goat with mediastinal lymph nodes CLA. The high occurrence of the virulent genes may indicate that these strains are highly virulent and they possess high virulence potential. C. pseudotuberculosis also showed sensitivity toward the majority of antibiotics used in this study. Phylogenetic tree of partial sequences of the rpoB gene showed that the biovar ovis isolates from both sheep and goat are grouped together in the same lineage and the nucleotide variation between the sequences does not change the protein expression. Thus confirming that the rpoB gene allows differentiation of C. pseudotuberculosis isolates at biotypes level. However, for the future larger number of the C. pseudotuberculosis isolates both visceral and superficial lymph nodes and from wider geographical areas can be included for better study of virulence potential and identification of C. pseudotuberculosis isolates at subspecies level using phylogeny based on rpoB.
CONFLICT OF INTEREST
There is no conflict of interest regarding the publication of this paper.
REFERENCES






