Advances in Animal and Veterinary Sciences
Mini Review
Advances in Animal and Veterinary Sciences. 2 (1): 50 – 55.Nucleic Acid Aptamers as an Emerging Diagnostic Tool for Animal Pathogens
Tarun Kumar Sharma1*, Ravi Shukla2*
2. Centre for Advanced Materials and Industrial ChemistrySchool of Applied SciencesHealth Innovation Research Institute RMIT University GPO Box 2476, Melbourne 3001 VIC Australia
*Corresponding author: [email protected], [email protected]
ARTICLE CITATION: Sharma TK and Shukla R (2014). Nucleic acid aptamers as an emerging diagnostic tool for animal pathogens. Adv. Anim. Vet. Sci. 2 (1): 50 – 55.
Received: 2013–12–12, Revised: 2013–12–25, Accepted: 2013–12–26
The electronic version of this article is the complete one and can be found online at (http://dx.doi.org/10.14737/journal.aavs/2014.2.1.50.55) which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Abstract
Various microbial and viral pathogens continuously posing threat to mankind and animals especially in agriculture economy based nations. In order to combat these challenges we need accurate diagnostic methods followed by proper treatment. Unfortunately contemporary detection methods are either time consuming or need sophisticated instruments so cannot be used for real field applications. Aptamer technology emerged as potential rival of antibody with several advantages over the later. Various aptamer have been generated against myriad of microbial and viral pathogens with the aim of detection and mitigation. In this review we tried to summarize the diagnostic application of aptamer technology for animal pathogens.
Introduction
Once Watson and Crick demonstrated the structure of DNA in 1953, it was thought that this molecule of life carry genetic information from one generation to next. But beside this, nucleic acids (DNA and RNA) have been found to perform some other interesting functions. DNA and RNA are often considered as huge molecular hard disk for information storage, but they might also have great potential to be used as diagnostic tool and therapeutic agents (Daniela et.al., 2005; Tuerk et.al., 1990;Wilson et.al., 1999; Regina et.al., 2007). Due to the extensive structural and conformational topology single–stranded (ss) polynucleotides (ssDNA and RNA) can fold into diverse conformations and directly interact with cellular proteins and other ligands. Such studies have opened a new research field leading to the selection and design of functional nucleic acid molecules that are known as “aptamers”. The name was derived from the Latin word “aptus”, meaning “to fit”, and these aptamers are agents to fit basically any given structure (Wilson et.al., 1999; Regina et.al., 2007). In 1990, Szostak and Gold independently pioneered an in vitro evolutionary process termed SELEX (Systematic Evolution of Ligands by Exponential Enrichment) to identify nucleic acids (aptamers) specific to organic dyes and T4 DNA polymerase (Daniela et.al., 2005). Since then, a variety of aptamers have been generated against myriad of targets. Moreover several attempts has been made to improve and to develop variant of this process to generate aptamers for specific purpose (Daniela et.al., 2005; Wilson et.al., 1999;. Beauty of this process lies in the fact that aptamers can be generated against any target ranging from simple small molecule to complex protein to whole cell or organism (Cancer cells, Bacteria, Protozoan etc.). conceptually aptamers are quite different from antisense nucleic acid molecules, which are considered linear molecules and block transcription according to the Watson–Crick base–pairing model. Aptamers are molecules with complex 3–dimensional (3–D) structures which function through affinity–binding to target molecules, in a fashion similar to antibody . Aptamers can fold into 3–D structures as a result of intramolecular interaction and bind to targets through a small number of contact points which determine high specificity. They are more stable than antibodies and can undergo denaturation and renaturation (Daniela et.al., 2005; Tuerk et.al., 1990; Wilson et.al., 1999; Regina et.al., 2007). Aptamers (8–15 kDa) are smaller than antibodies (155 kDa) and therefore have higher permeability and can penetrate targets more easily. Reporter molecules, such as fluorophores, biotin, or nanoparticles, can be easily attached to aptamers by chemical synthesis. Aptamers are derived from an in vitro evolutionary process, so toxins or molecules that do not elicit immune responses can also be used as target to develop aptamers. The in vitro process can also be performed in non–physiological selection conditions. Moreover, SELEX confers aptamers with high specificity and is able to discriminate targets of slight structural difference, such as isoforms of protein enzymes. Aptamer have typical dissociation constants ranging from the low picomolar to low nanomolar or sub micromolar (Wilson et.al., 1999; Regina et.al., 2007). A head to head comparison of aptamer and antibody is given in table 1. SELEX is an effective technology for the in vitro selection of aptamers that have high specificity to and affinity for a particular target. Many aptamers not only evinced high specificity and affinity, but also interfere with biological functions of target molecules, thus presenting themselves as prospective therapeutic candidates. Prof. Ellington has created a comprehensive aptamer database which is an online resource of all selected aptamer sequences that may have diagnostic or therapeutic utility. This database is updated on monthly basis and is publicly available free of cost at http://aptamer.icmb.utexas.edu/ (Jennifer et.al., 2004).
Detection of microbial pathogens is really critical as accurate diagnosis is key to right treatment and cure. Detection, identification and quantification of microbial pathogens are crucial for public health protection. Areas where detection of microbial pathogens is critical include clinical diagnosis, water and environmental analysis, food safety, and biodefense. Microbial culture–based tests and molecular assays (immunological or nucleic acid technologies) are the most common methodologies currently used (Lazcka et.al., 2007; Edith et.al., 2009). These techniques are either time consuming or require sophisticated equipment and highly trained personnel, hence increasing the analysis cost. A robust and rapid detection technique should provide dependable, real time, on–site, easy to use, and inexpensive detection with improved or equivalent sensitivity, specificity and reproducibility of culture–based tests (Edith et.al., 2009). According to Lazcka et al., biosensor technology is the fastest growing area in rapid diagnosis of microbial pathogens (Lazcka et.al., 2007). The commonly used biological recognition elements in biosensor platforms are antibodies and nucleic acid probes.
Recently, aptamers based detection emerged as platform technologies for variety of diagnostic application and they are being used alone or in combinations of quantum dots, modified and unmodified gold nanoparticles, aptamers based dot blot assays and ELISA as an electrical biosensor and also in combination with quartz crystal microbalance and surface enhance raman scattering (Edith et.al., 2009). In this review, an attempt has been made to summarize aptamer based detection methods for microbial pathogens of animal importance.
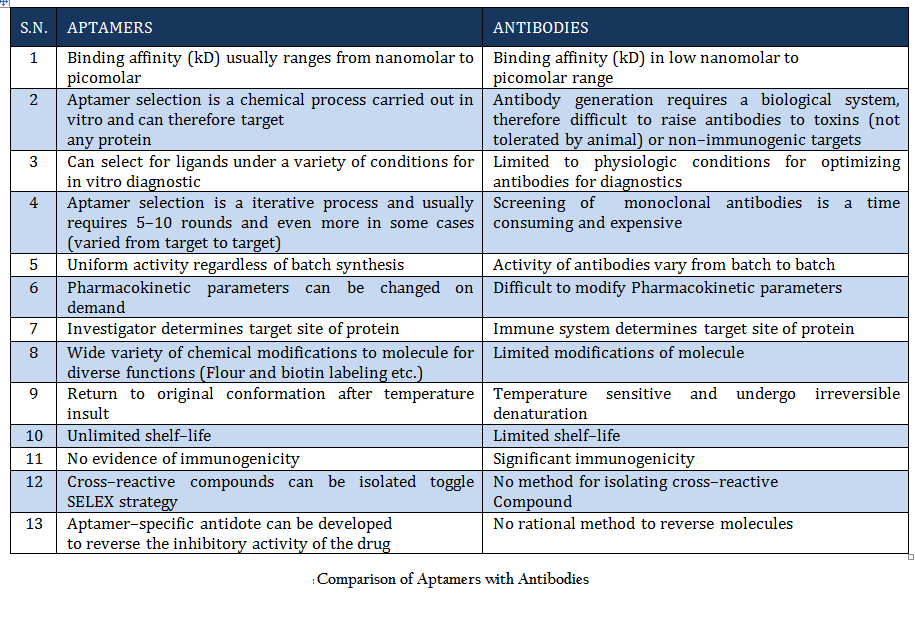
Comparison of Aptamers with Antibodies Aptamers are the rivals of Antibodies as they are stable, easy to develop and functionalized. More detail about aptamer advantages over antibody is given in Table 1.
SELEX; an Evolution in the Test Tube
SELEX is an evolutionary technique to identify a molecular key of interest (aptamer), from a huge bunch of keys (random sequence nucleic acid library), that has high specificity and affinity for its molecular lock (targets of interest). In the past decade, variety of aptamers has been generated against myriad of targets ranging from recombinant protein to small molecule to bacteria and viruses. In addition numerous modifications and variations have been made to enhance the versatility, specificity and affinity of this technique. In general, the SELEX process includes the following steps: the first step is to design a random, single–stranded DNA library. This library is chemically synthesized with a centralized random sequence (35–50 nucleotides long) flanked by fixed sequences at either end which served as primer binding domain during PCR (Daniela et.al., 2005; Tuerk et.al., 1990; Wilson et.al., 1999; Regina et.al., 2007). The second step is to incubate the random library with the target. Usually, small molecules and proteins are immobilized on solid supports to generate affinity matrices while in case of microbial pathogens either pathogen specific protein or whole cell of a particular pathogen is used as a target for aptamer selection. The third step is to separate bound oligonucleotides from unbound this is usually achieved by number of stringent washes. The fourth step is to separate bound nucleic acid from the target followed by enrichment of binders using polymerase chain reaction (PCR). Since PCR product is double ssDNA, therefore, in order to begin next round of aptamer selection one need to generate ssDNA this is normally achieved through asymmetric PCR, denaturing PAGE or using biotin–streptavidin interactions. The fifth and final step is to elute the ssDNA and being next round of selection. The use of RNA aptamer requires have to go two additional steps; one is in vitro RNA transcription and other is reverse transcription polymerase chain reaction. In order to enrich the binder’s population one has to go through these steps again and again. After reiterating these steps over several cycles (Figure 1), nucleic acids pool with high specificity to and affinity is cloned in appropriate cloning vector and subjected to sequencing. Then, binding affinity and specificity of nucleic acids with different motifs to target molecules can be tested and compared, and the aptamers with the highest affinity and specificity or other desired features is selected.
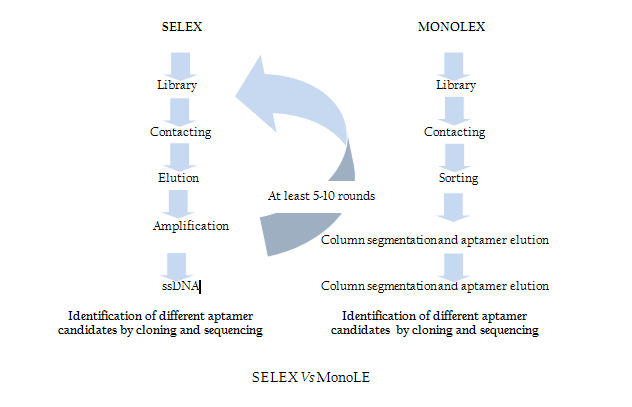
Modification of SELEX
Traditional SELEX protocol comprises iterative rounds of selection which is very time consuming and tedious job. In order to overcome this problem recently Andreas et al. introduced a single step MonoLEX process (Figure 2) of Aptamer selection (Nitsche et.al., 2007). This approach combined a single affinity chromatography step with subsequent physical segmentation of the affinity resin and one single final exponential amplification step of bound aptamers (Nitsche et.al., 2007). Therefore, this process improves the selection of high affinity aptamers by reducing the competition between the aptamers of different affinities during the PCR step.
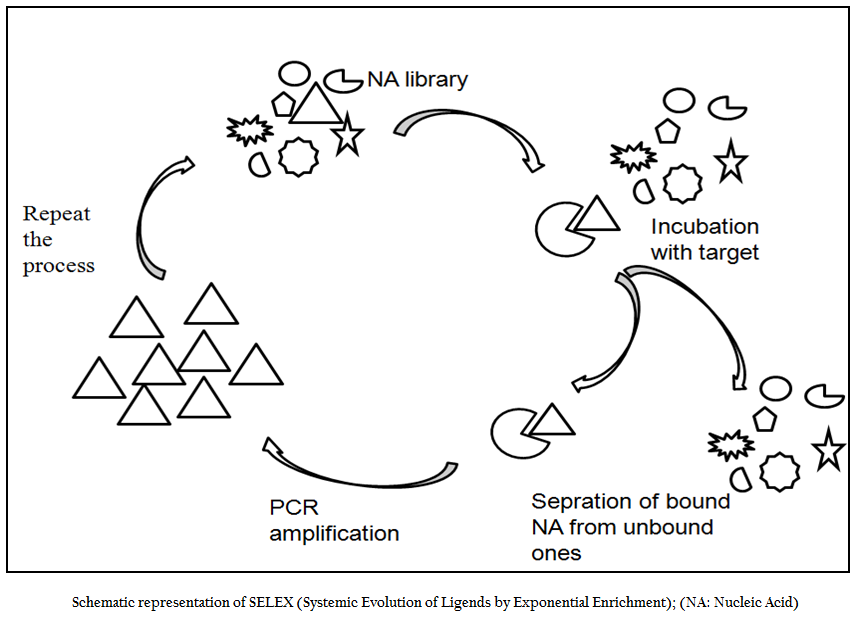
Figure 1:Schematic representation of SELEX (Systemic Evolution of Ligends by Exponential Enrichment); (NA: Nucleic Acid)
Aptamers against Bacterial Pathogens
Pathogenic diseases are among the most overwhelming burden across the world. They cause the suffering of hundreds of millions of people as well as an unknown figure of wild and domestic animals. Further the drugs present in the armory are very old and ineffective as pathogens have developed resistance against some of them. On the other hand process of developing new and improved anti–pathogenic drugs has been very slow. This is due to many factors, among them a significant lack of interest from the pharmaceutical industry as well as insufficient funding for drug discovery. Lastly, the drug discovery process involves screening of drug molecules from the huge library of synthetic compounds which is a slow and tedious process. Several attempts were made to generate aptamers against various bacterial pathogens which adversely affects animals and mankind.
DNA Aptamers Based Detection and Mitigation of Salmonellae
Salmonellae are one of the major causes of food borne sickness across the borders. It is responsible of about 1.4 million cases of salmonellosis including approximately 16,000 hospitalizations and 550 deaths only in United States (Joshi et.al., 2009). Salmonellae are also among bacterial pathogens in food animal species including cows (Wells et.al., 2001), pigs (Malorny et.al., 2005), chickens (Carli et.al., 2001), and turkeys (Nayak et.al., 2003). Amongst non–human cases, S. typhimurium is reported most frequently (Galanis et.al, 2006). Both classic zoonotic and food borne transmission of Salmonella spp. have been well documented for these animal species [Dejong et.al., 2005; Hendriksen et.al., 2004; Hsueh et.al. 2004; Schroeder et.al., 2005 Van et.al. 2004) . The organisms are shed in animal faeces periodically and at undetermined levels which makes its prevention and control a wild goose chase. Further detection of Salmonella and other bacterial pathogens in complex sample matrices such as feces, foods (Stevens et.al., 2004; Deisingh et.al., 2004), and environmental samples is really a challenging task because it needs sufficient enrichment in order to increase target copy number for downstream detection step. In addition, investigations at serovar level introduces complexity in diagnostic methodology. Though PCR can shorten the complexity but some time it fails or give false positive results when performed with complex biological samples (faces, food matrix etc.). In order to address such issues (Joshi et al., 2009) have demonstrated an aptamer based method for S.typhimurium diagnostics.
Recently (Olga et. al., 2013) generated ssDNA aptamers against S. enteritidis and S. typhimurium using cell–SELEX method. They have also demonstrated inhibitory potential of aptamers for Salmonella enteritidis and S. typhimurium. Interestingly, this study revealed when a bunch of aptamers were used together they evinced strong bacteriostatic action which could be attributed to the decrease in their membrane potential.
DNA Aptamers Based Detection of Francisella Tularensis
Vivekananda et al. generated ssDNA aptamers that bind to Francisella tularensis subspecies (subsp) japonica bacterial antigen (Vivekananda et.al., 2003). F. tularensis is an intracellular, nonmotile, nonsporulating, Gram–negative bacterial pathogen that causes tularemia in man and animals. Their group successfully isolated a set of 25 unique aptamer candidates that can specifically bind to F. tularensis subspecies japonica. When these aptamers were tested in a sandwich Aptamer–Linked Immobilized Sorbent Assay (ALISA) and dot blot format, the aptamer cocktail exhibited unique specificity in its ability to bind only to tularemia bacterial antigen from subspecies japonica, holarctica (also known as palaearctica) and tularensis but not to Bartonella henselae. Moreover, no cross reactivity was observed either to pure chicken albumin or chicken lysozyme. Thus, it appears that this novel antitularemia aptamer cocktail could be used as prospective diagnostic tool for a potential biological warfare agent like F. tularensis.
Aptamer against Virulent Mycobacterium Tuberculosis:
Tuberculosis (TB) remains a serious threat to mankind in developing nations. It is responsible for more than five million deaths per year worldwide. One–third of the world’s population is infected with Mycobacterium tuberculosis (MTB), the etiologic agent of TB. Global health problems attributed to TB need serious attention and in order to meet such challenges new and potent anti–TB drugs without cross–resistance with known antimycobacterial agents is urgently needed. (Chen et. al., 2007) generated first aptamers (ssDNA aptamer) NK2 using cell–SELEX against live MTB yielded aptamers that specifically recognize H37Rv over BCG. In addition, it modestly enhanced the survival rate of mice infected with the virulent strain H37Rv. Aptamer candidate (NK2) not only preferentially binds H37Rv but also increases the levels of interferon–γ in CD4+ T cells, and reduces bacterial counts in the spleens of infected mice (Chen et. al., 2007). Although neither the bacterial surface target nor the mechanism of protection has been identified, the authors proposed that NK2 most likely binds to and interferes with a self–protective H37Rv membrane protein, thus leading to more effective cell–mediated immunity. This strategy suggests aptamer NK2 can be used as diagnostic and therapeutic agent for MTB.
Chen et. al., 2009 have reported inhibitory action of aptamers NK2 for H37Rv by demonstrating that aptamer NK2 and 10th pool potentially inhibits H37Rv invasion to macrophages. Further their results showed H37Rv treated with NK2 or 10th pool significantly stimulated the secretion of interferon from macrophages as compared to control group.
Aptamer Based Detection of Foot and Mouth Disease Virus
Foot and Mouth Disease Virus (FMDV) is one of the serious threats to agriculture based economy of both developed and developing nations. It has caused several devastating outbreaks in the United States, United Kingdom and Asia including India. Due to such economic burden there is an urgent need to develop a rapid and reliable detection system for FMDV. Although several immunoassay and reverse transcription polymerase chain reaction (RT–PCR) and other nucleic acid amplification based methods are in place with wide range of FMD serotype specificity but unfortunately none of these methods is available in portable format or point of care test format.
Bruno et. al., 2008 developed a DNA aptamers against a 14 amino acid peptide from VP1 structural protein of FMDV this protein is highly conserved among 16 strains of ‘O’ serotype of FMDV. In this study they have utilized the high affinity and specificity of polyclonal aptamers to develop a novel FRET based assay that can confirm the presence of FMD within few minutes (Bruno et. al., 2008). This method could be adopted in portable format due to availability of hand held spectroflourimeter. They labelled VP1 structural peptide with Black Hole Quencher–2 (BHQ–2) dye and polyclonal aptamer population were labelled with Alexa Flour 546–14–dUTP by PCR and allowed to bind the BHQ–2 peptide conjugate. Following FRET aptamer–peptide complex, a light off or turn off kind response was observed within 10 minutes and this assay was able to detect as low as 25ng/ml VP1 peptide. Surprisingly, when individual aptamer candidate were used in similar kind of assay, they resulted in poor sensitivity and specificity which suggests that combinatorial action of aptamers leads to enhance sensitivity of assay. Further, unlike signalling aptamers (Jhaveri et.al., 2000) or aptamer beacons, (Hamaguchi et.al., 2001) these competitive FRET–aptamers attempt to place a known quencher target conjugate within the Förster distance of fluorophores in the aptamer–binding pocket and then compete it off with unlabeled analyte. In future, this approach could be used to develop point of care diagnostic assay for FMDV.
Selection of Aptamers against Live Trypanosomes
Among protozoan Trypnosomes are one of the serious threat to humans and cattle. It is the causative agent of sleeping sickness among cattle and humans. This disease is life threatening if remained undiagnosed and untreated. Moreover the available line of drugs causes serious adverse effects (side effects). In addition to that, development of drug resistance among trypanosomes is also a serious concern which has created a grave situation. Further, trypanosomes escape the immune response of invaded hosts through antigenic variation. This process is achieved through temporal expression of immunologically unrelated variants of surface glycoproteins (Blum et.al., 1993; Homann et.al., 1999). Fortunately, trypanosomes surface also contain some invariant proteins. In order to target such invariant surface protein Homann and Goringer designed SELEX experiment to generate RNA aptamers against invariant protein of live T. brucei with the aim to redirect the immune response to the surface of parasite using aptamers (Homann et.al., 1999). After 12 iterative cycles of selection and enrichment of binders, they were able to identify some pseudoknott aptamers that bind to blood stream stage trypanosomes and were not able to differentiate between two immunologically distinguishable T. brucei cell lines (MITat1.2 and MITat 1.4). Further they have also identified aptamer interaction partner be zero distance photo crosslinking and it was identified as 42kDa polypeptide. As expected this polypeptide was present on the surface of blood stream stage MITat1.2 and 1.4 but could not be identified on insect stage trypanosomes. Further experiments revealed that this 42kDa protein is a component of flageller pocket of the parasite. The same research group performed another study to determine fate of selected RNA aptamer (Homann et.al., 2001). They demonstrated when this aptamer binds to its target at flageller pocket it gets internalized and subsequently transported to lysosomes. This uptake pathway was visualized by flourphore labeled aptamer candidate. They further showed this intracellular internalization was sequence dependent as no such internalization was observed when truncated or random sequence was used. In addition to that, they have also reported that on internalization aptamer gets partially degraded to attain a stable 50 nucleotide structure. This work was sort of path breaking study which showed aptamer potential in mapping of surface associated protein to identify novel cell surface markers. Further, this work also suggests the application of aptamers as delivery vehicle to deliver aptamer–coupled compounds to lysosomal compartment.
CONCLUSION
Field of diagnostics always demand simple, rapid and reliable diagnostic methods that could also be adapted in point of care format for real field applications. Contemporary method of microbial pathogens relies on culture based, immunological and PCR based methods. All of these methods are tedious, time taking and require sophisticated instruments and skilled manpower. The field of aptamer technology opened a new vista in the field of diagnostics and therapeutics. Due to several advantages over antibody including negligible batch to batch variation these chemical rivals can easily be used to develop large scale point of care diagnostic assay. Further, ease in conjugation with gold nanoparticles (GNPs), quantum dots ribozymes and florophores makes them an attractive choice of diagnostic platform development. When these aptamer are conjugated with various signaling agents, they give visual readouts visible to naked eye. The enormous potential of aptamer remains to be exploited in the area of diagnostics.
ACKNOWLEDGEMENTS
We highly acknowledge the facility provided by our parent institutes (Translational Health Science and Technology Institute–India and RMIT University–Australia) to carry out this review work.
CONFLICT OF INTEREST
None
REFERENCES
Blum, M.L., Down, J.A., Gurnett, A.M., Carrington, M., Turner, M.J., Wiley, D.C., A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature (1993) 362:603–609.
http://dx.doi.org/10.1038/362603a0
PMid:8464512
Bruno J.G., Maria P. C., and Taylor P. Development of DNA Aptamers to a Foot–and–Mouth Disease Peptide for Competitive FRET–Based Detection J. Biomol. Tech. (2008) 19:109–115
PMid:19137093 PMCid:PMC2361164
Carli K.T., Unal C.B., Caner V., Eyigor A., Detection Salmonellae in chicken feces by a combination of tetrathionate broth enrichment, capillary PCR, and capillary gel electrophoresis. J Clin Microbiol (2001) 39:1871–1876.
http://dx.doi.org/10.1128/JCM.39.5.1871-1876.2001
PMid:11326006 PMCid:PMC88041
Chen. F. et al. (2012) Aptamer inhibits Mycobacterium tuberculosis (H37Rv) invasion of macrophage Mol Biol Rep (2012)39:2157–2162
http://dx.doi.org/10.1007/s11033-011-0963-3
PMid:21643749
Chen. F. et al. Development of DNA aptamers as thereaupetic agent against Mycobacterium tuberculosis. Biochem. Biophy. Res. Commun. (2007) 357:743–748
Daniela P., Michael B., Raymund B., Ansgar R., Aptamers—basic research,drug development,and clinical applications. Appl Microbiol Biotechnol (2005) 69:367–374
http://dx.doi.org/10.1007/s00253-005-0193-5
PMid:16283295
De Jong B., Andersson Y., Ekdahl K. Effect of regulation and education on reptileassociatedsalmonellosis. Emerg Infect Dis (2005) 11:398–403.
http://dx.doi.org/10.3201/eid1103.040694
PMid:15757554 PMCid:PMC3298264
Deisingh A.K, Thompson M., Biosensors for the detection of bacteria. Can J Microbiol. (2004)50:69–77.
http://dx.doi.org/10.1139/w03-095
PMid:15052308
Edith T. C., Evangelyn C. A., Aptasensors for detection of microbial and viral pathogens . Biosens Bioelectron (2009) 24:3175–3182
http://dx.doi.org/10.1016/j.bios.2008.11.010
PMid:19117748
Galanis E., Lo Fo Wong D. M., Patrick M.E., Binsztein N., Cieslik A., Chalermchikit T., et al. Web–based surveillance and global Salmonella distribution, 2000–2002. Emerg Infect Dis (2006)12:381–388.
http://dx.doi.org/10.3201/eid1203.050854
http://dx.doi.org/10.3201/eid1205.050854
PMid:16704773 PMCid:PMC3291443
Hamaguchi N., Ellington A., Stanton M. Aptamer beacons for the direct detection of proteins. Anal Biochem (200?????)1294:126–131.
Hendriksen S.W., Orsel K., Wagenaar J.A., Miko A., van Duijkeren E., Animal–tohuman transmission of Salmonella typhimurium DT104A variant. Emerg Infect Dis (2004)10:2225–2227.
http://dx.doi.org/10.3201/eid1012.040286
PMid:15663868 PMCid:PMC3323380Homann, M., Go¨ringer, H.U., Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. (1999) 27: 2006–2014.
http://dx.doi.org/10.1093/nar/27.9.2006
PMid:10198434 PMCid:PMC148414
Homann, M., Go¨ringer, H.U., Uptake and intracellular transport of RNA aptamers in African trypanosomes suggest therapeutic 'piggyback' approach. Bioorg. Med. Chem. (2001) 9, 2571–2580.
http://dx.doi.org/10.1016/S0968-0896(01)00032-3
Hsueh P.R., Teng L.J., Tseng S.P., Chang C.F., Wan J.H., Yan J.J., Ciprofloxacin resistant Salmonella enterica typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerg Infect Dis (2004)10:60–68.
http://dx.doi.org/10.3201/eid1001.030171
PMid:15078598 PMCid:PMC3322755
Jennifer F. L., Jay R.H., Lauren A. M., Andrew D. E., Aptamer Database. Nucleic Acids Res. (2004) 32:95–100
http://dx.doi.org/10.1093/nar/gkh094
PMid:14681367 PMCid:PMC308828
Jhaveri S., Rajendran M., Ellington A., In vitro selection of signalling aptamers. Nature Biotechnol (2000)18:1293–1297.
http://dx.doi.org/10.1038/82414
PMid:11101810
Joshi R., Janagama H., Dwivedi H. P., Senthil Kumar T.M.A., Jaykus L. A., Schefers J., Sreevatsan S., Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol. Cell. Probes (2009) 23: 20–28
http://dx.doi.org/10.1016/j.mcp.2008.10.006
PMid:19049862
Lazcka O., Del F.J., Munoz F.X., Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron (2007) 22:1205–1217.
http://dx.doi.org/10.1016/j.bios.2006.06.036
PMid:16934970
Malorny B., Hoorfar J., Toward standardization of diagnostic PCR testing of fecalsamples: lessons from the detection of Salmonellae in pigs. J Clin Microbiol(2005) 43:3033–3037.
http://dx.doi.org/10.1128/JCM.43.7.3033-3037.2005
PMid:16000411 PMCid:PMC1169119
Nayak R., Kenney P.B., Keswani J., Ritz C., Isolation and characterisation of Salmonella in a turkey production facility. Br Poult Sci (2003) 44:192–202.
http://dx.doi.org/10.1080/0007166031000088370
PMid:12828204
Nitsche A., Kurth A., Dunkhorst A., Pänke O., Sielaff H., Junge W., Muth D., Scheller F., Stöcklein W., Dahmen C., Pauli G., Kage A. One–step selection of Vaccinia virus–binding DNA aptamers by MonoLEX. BMC Biotechnol. (2007) Aug 15:43:48
Olga S. K., Anna G. S., Tatiana N. Z., Irina T.R., Galina S., Z., Evgeny N. E., Xiaoyan W., Mohamed W., Alla B. S., Olga V. P., Olga A. Z., Ekaterina A. S., Vasily S. M., Yury E. G., Nadezhda M. T., Maxim V. B., Anna S. Z., Development of Bacteriostatic DNA Aptamers for Salmonella. J. Med. Chem. (2013) 56:1564−1572
Regina S., Christine R., Beate S., SELEX—A (r)evolutionary method to generate high–affinity nucleic acid ligands. Biomol. Eng. (2007) 24:381–403
http://dx.doi.org/10.1016/j.bioeng.2007.06.001
PMid:17627883
Schroeder C. M., Naugle A.L., Schlosser W.D, Hogue A.T, Angulo F.J, Rose J.S., Estimate of illnesses from Salmonella enteritidis in eggs, United States. Emerg Infect Dis (2005)11:113–115.
http://dx.doi.org/10.3201/eid1101.040401
PMid:15705332 PMCid:PMC3294346
Stevens K.A., Jaykus L.A., Bacterial separation and concentration from complex sample matrices: a review. Crit Rev Microbiol (2004)30:7–24.
http://dx.doi.org/10.1080/10408410490266410
PMid:15116760
Tuerk C., Gold L., Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science (1990) 249:505–510.
http://dx.doi.org/10.1126/science.2200121
PMid:2200121
Van Immerseel F., Pasmans F., De Buck J., Rychlik I., Hradecka H., Collard J.M., Cats as a risk for transmission of antimicrobial drug–resistant Salmonella. Emerg Infect Dis (2004)10:2169–2174.
http://dx.doi.org/10.3201/eid1012.040904
PMid:15663855 PMCid:PMC3323385
Vivekananda J., Kiel J.L., Methods and components for aptamers against anthrax. Patent 6,5696,30 B1 issued on May 27, 2003.
Wells S. J., Fedorka–Cray P. J., Dargatz D.A., Ferris K., Green A., Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J Food Prot (2001) 64:3–11.
PMid:11198437
Wilson D.S., Szostak, J.W., In vitro selection of functional nucleic acids. Annu. Rev. Biochem (1999) 68:611–647.
http://dx.doi.org/10.1146/annurev.biochem.68.1.611
PMid:10872462
Featuring
-
Effect of Dietary Selenium Conjugated Protein Black Soldier Fly Larvae Meal on Carcass Characteristics and Meat Fatty Acid Profiles in Broiler Duck
David Kurniawan, Eko Widodo, Agus Susilo, Osfar Sjofjan
Adv. Anim. Vet. Sci., Vol. 13, Iss. 1, pp. 108-114
-
The Relevance of Two-Doses of Foot-And-Mouth Disease (FMD) Virus Vaccine on Immune Response in Beef Cattle
Esdinawan Carakantara Satrija, Sri Murtini, I Wayan Teguh Wibawan, Okti Nadia Poetri
Adv. Anim. Vet. Sci., Vol. 13, Iss. 1, pp. 103-107
-
Physiological and Histological Consequences of Growth Stunting in Broiler Chickens
Qutaiba J. Gheni, Alfred S. Karomy, Alice Louis Yousaf, Wessam Monther Mohammed Saleh
Adv. Anim. Vet. Sci., Vol. 13, Iss. 1, pp. 96-102
-
Study of Noni Fruit Extract Microcapsule on Enzymes Activity, Digestibility and Hematologic of Sentul Chicken
Tuti Widjastuti, Wiwin Tanwiriah, Eka Wulandari, Sekarupa Rengganis Nusantara
Adv. Anim. Vet. Sci., Vol. 13, Iss. 1, pp. 89-95
Subscribe Today
Receive free updates on new articles, opportunities and benefits

© 2024 ResearchersLinks. All rights Reserved. ResearchersLinks is a member of CrossRef, CrossMark, iThenticate.