Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences. 2(1): 42 – 45Molecular Characterization of E. coli Isolated from Raw Vegetable
Abha Dutta, Namita Joshi*, Rajesh Kumar Joshi1, Akhilesh Kamal2 2
2. Department of Veterinary Microbiology / Department of Animal Husbandry (Govt. of India)
*Corresponding author: [email protected]
ARTICLE CITATION: Dutta A, Joshi N, Joshi PK and Kamal A (2014). Molecular characterization of e. coli isolated from raw vegetable. Adv. Anim. Vet. Sci. 2 (1): 42 – 45.
Received:2013–11–10 Revised:2013–12–15, Accepted: 2013–12–16
The electronic version of this article is the complete one and can be found online at (http://dx.doi.org/10.14737/journal.aavs/2014.2.1.42.45) which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Abstract
Vegetables that are commonly consumed in raw form, collected from retail shops viz. coriander leaves, radish, green chilies, cucumber and tomato were tested for contamination with E. coli. Out of 80 samples processed, 43 (53.75) percent samples yielded E. coli. The highest prevalence of E . coli i.e. 86.66 percent was observed in coriander samples followed by 60.00 % in tomato, 53.33 % in reddish, 50.00 %in green chili and 20 % in cucumber samples. The isolates belonged to serogroups O4, O5, O11, O17, O20, O60, O63, O65, O69, O71, O116, O132, O138, O147, three rough strains and 10 untypable. The isolates were subjected to PCR for detection of mdh, elt, est, stx1 and stx2 genes. The mdh gene was amplified in all 43 isolates while elt gene was detected in 30 isolates belonging to six different serogroups viz. O17, O60, O69, O138 and O147 and 7 untypable E. coli. The est gene was exhibited by 20 isolates belonging to five different serogroups viz. O17, O 60, O69, O138 and 3 rough strain and 5 untypable isolates. Amplification of stx1 gene was detected in only two isolates, both of O65 serogroup whereas, stx2 gene was observed in 9 isolates belonging to serogroup O69 and 3 rough strains.
Introduction
Use of animal manure for cultivation is a common practice and it is still considered to be better and safer than chemical fertilizers in India. Recently, association between fresh vegetables and outbreaks of food borne infections has led to a greater concern about contamination of vegetables with fecal pathogenic bacteria like E. coli, Vibrio, Salmonella, norovirus, Shigella, Listeria etc (Mahima et al., 2013) and safety of using animal manures as fertilizer in vegetable production (Pell, 1997; Tauxe et al., 1997). The animal faeces and manure contains number of bacteria that are zoonotic, such as Escherichia coli, Campylobacter spp. Salmonella spp. and Mycobacterium spp. (McGarvey et al., 2004; Dhama et al., 2013; Malik et al., 2013; Singh et al., 2013). Therefore, it is imperative to test the vegetables for indicator bacteria that are always present in bovine manure and have survival characteristics comparable to those of fecal enteric pathogens. The E. coli is one of the important enteric pathogens among the intestinal microflora of human and warm–blooded mammals. Hence, the present study was designed to study E. coli contamination of vegetables that are generally consumed raw or served as salad on dining table using conventional and molecular methods.
MATERIALS AND METHODS
Collection and Preparation of Samples
The study was conducted on the vegetables that are generally consumed raw as salad. Total of 80 samples consisting 15 each of tomato, cucumber, radish and coriander leaves and 20 samples of green chilies were collected from the local retail shops. The samples were collected in sterile polybags and brought to laboratory. In laboratory, the samples were washed with sterile PBS under sterile conditions and the washings were allowed to stand for ten minutes. The supernatant was discarded; the sediment was collected in sterile glass vials and stored at 4ºC until processed for isolation of E. coli.
Isolation and Identification of E. coli
The enrichment was done by inoculating a loopful of the sediment into MacConkey lactose broth at 370C for 18hr and then streaked on MacConkey Lactose Agar (MLA). Following 24 hr of incubation at 370C, the lactose fermenting rose–pink colonies were picked up and streaked onto Eosin Methylene Blue (EMB) agar plates. The samples producing a greenish metallic sheen on EMB plates after 24 hr incubation at 37ºC, were transferred on to nutrient agar slants and further processed for biochemical identification as described by Cruickshank et al. (1975). The isolates were identified on the basis of their cultural, morphological and biochemical characters (Edwards and Ewing, 1972) and those preliminarily screened out as E. coli were serotyped at National Salmonella and Escherichia Center, Central Research Institute, Kasauli, H.P., India.
Preparation of DNA Template
The snap chill technique described byIbenyassine et al. (2007) was used for preparation of DNA template for amplification of virulence genes. For this, 2ml nutrient broth culture of the field isolates was centrifuged at 4,000 rpm for 15 min. The supernatant was discarded; the pellet was suspended in 1ml of sterile triple glass distilled water (TGDW) and centrifuged further for 15min. The resultant pellet was suspended in 1 ml of TE buffer (pH 8.0) and centrifuged at 9,000 rpm for 6 min. The pellet so obtained was re–suspended in 500 µl of sterile TGDW, boiled for 10 min in a boiling water bath and then rapidly chilled at –20ºC. The supernatant was directly used as a template for further amplification of target sequence.
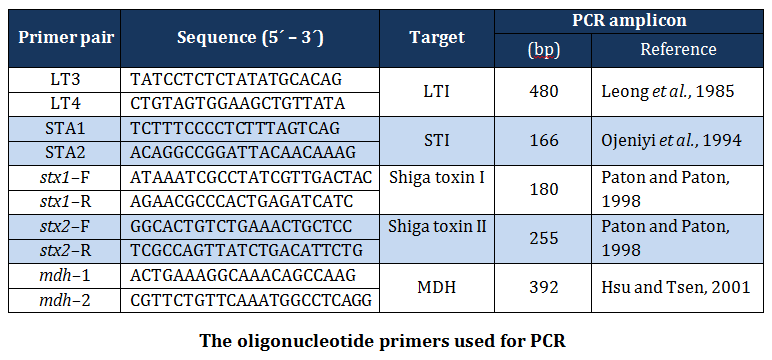
Primers
The published primers (Table 1) based on specific portion of mdh, elt, est, stx–1 and stx–2 genes of E. coli, were synthesized by Merk (India) and used in the present study.
The simplex PCR assays were carried out in 50µl of reaction mixture containing 25µl of 2X master mix (Merck), 5µl of DNA template, 10 picomoles of forward and reverse primers.
A negative control without template DNA was included in each experiment. PCR cycling was performed in a thermal cycler (Genie TC– 3000) at initial denaturation for 3 min at 94ºC, followed by 35 cycles of 94ºC for 20 sec., annealing at 60ºC for 30 sec., extension at 72ºC for 30 sec and final extension at 72ºC for 5 minutes. The amplified product was electrophoresed on a 1.5% agarose gel in Tris–Borate–EDTA buffer at 80Volts. The gels were stained with ethidium bromide and the bands were visualized under UV gel doc system (UVtech, UK).
RRESULT AND DISCUSSION
RESULT AND DISCUSSION
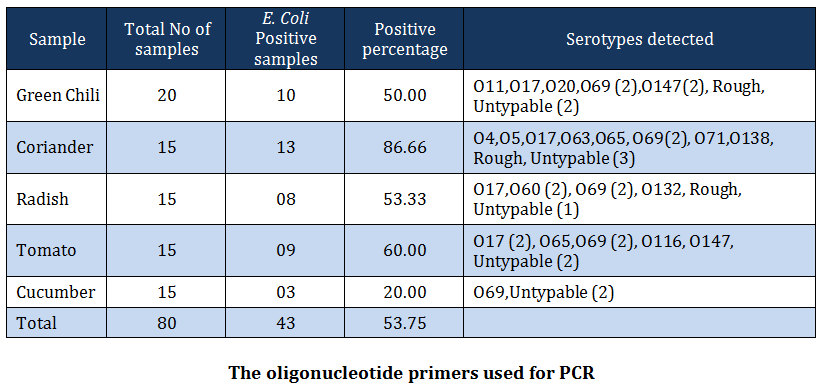
Out of 80 vegetable samples processed, 43 (53.75 percent) samples yielded E. coli. The prevalence of E. coli was recorded greatest in coriander samples (86.66 percent) followed by tomatoes (60.00 percent), radish (53.33 percent), green chilis (50.00 percent) and cucumber (20 percent) (Table –2). The distribution of various serotypes among vegetables tested is given in Table –2. The prevalence of E. coli in vegetable samples has been reported earlier by many workers (WHO, 1996; CDR, 1997; Buck and Walcott, 2003, Ibenyassine et al., 2007). There are reports that provide the evidence that E. coli could be transmitted to vegetables from manure (Wang and Doyle 1998; Natvig et al., 2002), contaminated irrigation water (Fattal et al., 1986; Rai and Tripathi, 2007) and direct contamination from animal and human faeces (Johannesen et al., 2002). Solomon et al. (2002) also reported the survival of E. coli O157:H7 on lettuce leaves for more than 20 days and concluded that the organism migrated to the internal location in plant tissue and was protected from action of sanitizing agent.
In the present study, E. coli isolates of serogroup O4, O5, O11, O17, O20, O60, O63, O69, O71, O116, O132, O138 and O147 were recovered from vegetable washings. Of these, O69 and O147 serogroups were also reported from cases of bovine diarrhea (Diwakar et al., 2008) and serogroups O17, O60, O69, O147 from poultry (Dwivedi et al., 2007; Yadav et al., 2009) from the same study area. Similarly, serogorup O5, O20, O60 and O138 have been reported from diarrheic pigs by Dutta et al. (2010). These findings clearly indicate the association of manure with the isolation of E. coli from vegetables. Besides this, serogroups O4, O20, O60 and O132 have been reported to be associated with human cases of urinary tract infections, pyelonephritis and renal failure (Johnson, 1991; Blanco et al., 1997; Johnson et al., 2005; Kausar et al., 2009). This indicates that the vegetables sold in domestic market pose serious public health risk as these are injured while peeling, slicing, shredding during salad preparation and usually consumed without heat treatment (Beuchat, 2002). Furthermore, the predominance of O69 serogroup in this study warrants a special attention to study their role in the development of diarrhea because serogroup O69 is an established shiga toxin producing E. coli strain (Griffin and Tauxe, 1991). Serogroup O20 obtained in this study is a well – known human enteric pathogen and has been isolated most frequently from the water in Coimbatore (Grover, 2006).
Conventional culture techniques are universally recognized as gold standard method for diagnosis of E. coli but this process is rather lengthy and may last 5–10 days or more. Many foods with short shelf lives would have been consumed by the time the result of analysis becomes available. Hence, the development of simple and rapid assays would enable identification of E. coli in contaminated foodstuffs in a timelier manner. Therefore, an attempt was made to detect E. coli in vegetable washings using E. coli mdh gene primer with minimum amount of time. The PCR amplification of mdh gene gave positive result in all 43 isolates and this was in conformity with the findings of Ibenyassine et al. (2007). Hsu and Tsen (2001) also used mdh gene for direct detection of E. coli from milk and water samples. The enzyme malate dehydrogenase from E. coli catalyzes the interconversion of malate and oxaloacetate, a critical step in carbohydrate metabolism (Wright and Viola, 2001) and thus, detection of its gene can specifically be used for direct detection of E. coli in vegetable washings (Ibenyassine et al., 2007).
The differentiation of E. coli pathotypes requires detection of virulence genes either by biological assays or by molecular techniques. Numerous virulence genes were also targeted in present study by simple PCR assay. The presence of elt gene was detected in 30 isolates of E. coli that belonged to the serogroup O17 (5 strains), O60 (2 strains), O69 (9 strains), O138 (1 straian), O147 (3 strains), 3 rough strains and all ten untypable. The PCR amplification of est gene gave positive results in serogroups O17 (5 strains), O60 (2 strains), O 69 (9 strains), O138 (1 strain), 3 rough strains and all ten untypable
E. coli. In a study, Toma et al. (2003) compared the efficacy of simplex and multiplex PCR to categories ETEC strains by targeting elt and est genes. The present study indicated that ETEC strains belonged to the serogroup O26, O27, O126 and O128. Both the genes were also detected in six untypable strains which were in conformation with our findings. Toni et al. (2006) reported presence of elt gene in O149 and est gene in O8, O20, O64, O101 and O149 serogroups. Presence of these genes indicate the enterotoxigenic nature of these isolates and such ETEC isolates were reported to have the ability to cause profuse, watery diarrhea by release of LT, ST or both enterotoxins (Nataro and Kaper, 1998).
In India, isolation of STEC has been reported from cattle (Pal et al., 1999), sheep (Wani et al., 2004; Bhat et al., 2008), fish (Kumar et al., 2001), human faeces (Khan and Ner, 2002) and piglets (Dutta et al., 2010). But the information related to the prevalence of STEC in vegetables is rare. Hence, in the present study, the isolates were screened for presence of stx genes to establish their STEC nature. The stx1 gene was detected in two isolates both of serogroup O65, however, stx2 gene was demonstrated in 12 isolates that were representative of O69 (9) and rough strains (3). None of the isolates showed the amplification of both stx1 and stx2 genes. However, Dutta et al. (2010) reported the presence of stx1 as well as stx2 gene in O60 and O138 serogroups isolated from an organized pig farm in Kolkata. Toma et al. (2003) have reported stx gene in the serogroups O15, O28, O111, O121, O145 and O157 and placed them in STEC category. The STEC strains have been characterized by production of either one or both of the two major types of Stx proteins designated as shiga like toxin–1 (stx1toxin) and shiga like toxin–2 (stx2 toxin) (Rangel and Sparling, 2005) and the production of the later is associated with increased risk of developing Haemolytic Uraemic Syndrome (Boerlin et al., 1999). STEC strains have also been isolated from a number of environmental sources (Rangel and Sparling, 2005) that may result in contamination of vegetables. Detection of stx1 and stx2 genes only in two and nine isolates, respectively in present study may also be attributed to loss of virulence genes during subculture as reported by Karch et al. (1992).
REFERENCES
Beuchat LR (2002). Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. J Microb Infect 4: 413–423.
http://dx.doi.org/10.1016/S1286-4579(02)01555-1
Bhat MA, Nishikawa Y and Wani SA (2008). Prevalence and virulence gene profiles of shiga toxin producing Escherichia coli and enteropathogenic Escherichia coli from diarrheic and healthy lambs in India. Small Ruminant Res 75: 65–70.
http://dx.doi.org/10.1016/j.smallrumres.2007.08.006
Blanco M, Blanco JE, Alonso MP, Mora A, Balsalobre C and Munoa F (1997). Detection of pap, prs & afa adhesin–encoding operons in uropathogenic E.coli strains: relationship with expression of adhesions and production of toxin. Res Microbiol 148:745–55.
http://dx.doi.org/10.1016/S0923-2508(97)82450-3
Boerlin P, McEwan SA, Boerlin–Petzold F, Wilson JB, Johnson RP and Gyles CL (1999). Association between virulence factors of Shiga toxin producing Escherichia coli and diseases in humans. J Clin Microbiol 37: 497–503.
PMid:9986802 PMCid:PMC84443
Buck J.W. and Walcott R.R. (2003). Recent trends in microbiological safety of fruits and vegetables. Plant Health Progress. 10: 1092–1098.
CDR(1997). Hospital outbreak of E. coli O157:H7 associated with a rare phage type, Ontario. Canada Communicable Disease Report 23: 04–05
Cruickshank R, Duguid JP, Marmion BP and Ewing WH (1975). Identification of Enterobacteriacae 3rd ed. Burges publishing Co. Atlanta, Georgia, U.S.A., pp 152–154.
PMid:1182790
Dhama, K., Rajagunalan, S., Chakraborty, S., Verma, A.K., Kumar, A. and Tiwari, R. and Kapoor, S. (2013). Food–borne pathogens of Animal origin–Diagnosis, prevention and control and their zoonotic significance– A review. Pak. J. Biol. Sci. 16(20): 1076–1085.
http://dx.doi.org/10.3923/pjbs.2013.1076.1085
PMid:24506006
Diwakar RP, Joshi N, Joshi RK, Niyogi D and Dwivedi K (2008). Serotyping and Antibiogram of E.Coli isolated from Diarrhoeic calves. Indian J Anim Hlth 47: 119–121.
Dutta TK, Roychoudhury P, Sadhukhan T K, Ghosh A and Bandyopadhaya AG (2010). Detection and characterization of shiga toxigenic E. coli (STEC) and enteropathogenic Escherichia coli (EPEC) from diarrhoeic piglets in an organized farm in Kolkata. Indian J Anim Sci 80: 493–496.
Dwived K, Joshi RK, Joshi N, Diwakar RP and Niyogi, D. (2007). Isolation and Antibiogram of E.coli from Poultry in Purvanchal region of Uttar Pradesh. Indian J Anim Hlth 46: 77–80.
Edwards PR and Ewing WH (1972). Identification of Enterobacteriaceae, 3rd edn. Burges Publishing Co. Atlanta, Georgia, U.S.A.
Fattal B, Wax Y, Davies M and Shuval HI (1986). Health risk associated with wastewater irrigation. American J Pub Hlth 76:977–9.
http://dx.doi.org/10.2105/AJPH.76.8.977
Griffin PM and Tauxe RV (1991). The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E.coli and the associated hemolytic uremic syndrome. Epidemiological Rev 13: 60–98.
PMid:1765120
Grover YP (2006). Virulence factors associated with avian pathogenic E. coli. Cur Meth Vet Microbiol pp 116–120.
Hsu S, Tsen H (2001). PCR primer designed from malic acid dehydrogenase gene and their use for detection of Escherichia coli in water and milk samples. Int J Food Microbiol 64: 1–11.
http://dx.doi.org/10.1016/S0168-1605(00)00425-6
Ibenyassine K, Jaa A, Yahayan K and Moulay ME (2007). A simple and rapid detection by PCR of enteropathogenic Escherichia coli in naturally contaminated vegetables. J Rapid Meth Auto Microbiol 16 :113–121.
http://dx.doi.org/10.1111/j.1745-4581.2008.00110.x
Johannesen GS, Loncarevic S and Kruse H (2002). Bacteriological analysis of fresh produce in Norway. Int J Food Microbiol 77:199–204.
http://dx.doi.org/10.1016/S0168-1605(02)00051-X
Johnson JR (1991). Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4:80–128.
PMid:1672263 PMCid:PMC358180
Johnson JR, Owens K, Gajewski A and Kuskowski MA (2005). Bacterial characteristics in relation to clinical source of Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women., J Clin Microbiol 43:6064–6072.
http://dx.doi.org/10.1128/JCM.43.12.6064-6072.2005
PMid:16333100 PMCid:PMC1317206
Karch H, Meyer T, Russmann H and Heesemann J (1992). Frequent loss of Shiga–like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun 60: 3464–3467.
PMid:1639518 PMCid:PMC257340
Kausar Y, Chunchanur SK, Nadagir LH, Halesh MSD and Chandrasekhar R (2009). Virulence factors, Serotypes and Antimicrobial Suspectibility Pattern of Escherichia coli in Urinary Tract Infections. Al Ameen J Med Sci 2:47 –51.
Khan MA and Ner TS (2002). Mechanisms of emerging diarrheagenic Escherichia coli infection. Curr Infect Dis Report 4: 112–7.
http://dx.doi.org/10.1007/s11908-002-0050-y
PMid:11927041
Kumar SH, Otta SK and Karunasagar I, (2001). Detection of Shiga–toxigenic Escherichia coli (STEC) in fresh seafood and meat marketed in Mangalore, India. Appl Microbiol 33: 334–38.
http://dx.doi.org/10.1046/j.1472-765X.2001.01007.x
Leong J, Vinal AC and Dallas WS (1985). Nucleotide sequence comparison between heat–labile toxin B–subunit cistrons from Escherichia coli of human and porcine origin. Infect Immun 48: 73–77.
PMid:3884513 PMCid:PMC261916
Leong, J. M., Fournier, R. S. and Isberg, R. R. (1990). Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO J., 9:1979–1989.
PMid:1693333 PMCid:PMC551907
Mahima, Verma, A.K, Tiwari, R., Karthik, K., Chakraborty, S., Deb, R. and Dhama, K. (2013). Nutraceuticals from fruits and vegetables at a glance: A review. Journal of Biological Sciences, 13(2): 38–47.
http://dx.doi.org/10.3923/jbs.2013.38.47
Mailk, S., Kumar, A., Verma, A.K., Gupta, M.K., Sharma, S.D., Sharma, A.K. and Rahal, A. (2013). Incidence and Drug Resistance Pattern of Collibacillosis in Cattle and Buffalo Calves in Western Uttar Pradesh in India. J. Anim. Health Prod. 1(1): 15–19.
McGarvey JA, Miller WG, Sanchez S and Stanker L (2004). Identification of Bacterial Populations in Dairy Wastewaters by Use of 16S rRNA Gene Sequences and Other Genetic Markers. App Environ Microbiol 70: 4267–4275.
http://dx.doi.org/10.1128/AEM.70.7.4267-4275.2004
PMid:15240310 PMCid:PMC444815
Nataro JP and Kaper JB (1998). Diarrheagenic Escherichia coli. Clin Microbiol Rev 11: 142–201.
PMid:9457432 PMCid:PMC121379
Natvig EE, Ingham SC, Ingham BH, Cooperband LR and Roper TR (2002). Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soil with incorporated bovine manure. Appl Environ Microbiol 68:2737–2744.
http://dx.doi.org/10.1128/AEM.68.6.2737-2744.2002
PMid:12039728 PMCid:PMC123957
Ojeniyi B, Ahrens P and Meyling A (1994). Detection of fimbrial and toxin genes in Escherichia coli and their prevalence in piglets with diarrhea. The application of colony hybridization assays, polymerase chain reaction and phenotypic assays. J Vet Med 41:49–59.
http://dx.doi.org/10.1111/j.1439-0450.1994.tb00205.x
Pal A, Ghosh S, Ramamurthy T, Yamasaki S, Tsukamoto T, Bhattacharya SK, Nair GB and Takeda Y (1999). Shiga toxin producing Escherichia coli from healthy cattle in a semi urban community in Calcutta, India. Indian J. of Med. Res. 110: 83–85.
PMid:10612908
Paton JC and Paton AW (1998). Pathogenesis and diagnosis of Shiga toxin–producing Escherichia coli infections. Clin. Microbiol Rev 11: 450–479.
PMid:9665978 PMCid:PMC88891
Pell AN (1997). Manure and microbes: public and animal health problem. J Dairy Sci 80: 2673–2681.
http://dx.doi.org/10.3168/jds.S0022-0302(97)76227-1
Rai PK and Tripathi BD (2007). Microbial contamination in vegetables due to irrigation with partially treated municipal waste water in tropical city. Iunter J Environ Health Res., 17: 385–395.
http://dx.doi.org/10.1080/09603120701628743
PMid:17924267
Rangel J and Sparling P (2005). Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis 11:603–609.
http://dx.doi.org/10.3201/eid1104.040739
PMid:15829201 PMCid:PMC3320345
Singh, S.V., Singh, A.V., Kumar, A., Singh, P.K., Deb, R., Verma, A.K., Kumar, A., Tiwari, R., Chakraborty, S. and Dhama, K. (2013). Survival mechanisms of Mycobacterium avium subspecies paratuberculosis within host species and in the environment – A review, Natural Science, 5(6): 710–723
http://dx.doi.org/10.4236/ns.2013.56088
Solomon EB, Yarin S and Metthews KR (2002). Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subseqent internalization. App Environ Microbiol 68: 397.
http://dx.doi.org/10.1128/AEM.68.1.397-400.2002
PMid:11772650 PMCid:PMC126537
Stevens L and Hobson K. (2011). Europe's E. coli Cases Rise. Wall Street J., Health Industry, JUNE 4, 2011
Tauxe R, Kruse H, Hedberg C, Potter M, Madden J and Wachsmuth K (1997). Microbial hazards and emerging issues associated with produce: a preliminary report to the National Advisory Committee on Microbiological Criteria for Foods. J Food Prot 60: 1400–1408.
Toma C, Lu Y, Higa N, Nakasone N, Chinen I, Baschkier A, Rivas M and Iwanaga M (2003). Multiplex PCR Assay for identification of human diarrheagenic Escherichia coli. J Clin Microbiol 41: 2669–2671.
http://dx.doi.org/10.1128/JCM.41.6.2669-2671.2003
PMid:12791900 PMCid:PMC156568
Toni A, Chapman Xi–Yang Wu, Idris Barchia, Karl A, Bettelheim Steven D (2006). Comparison of Virulence Gene Profiles of Escherichia coliStrains Isolated from Healthy and Diarrheic Swine. App Environ Microbiol 72: 4782–4795.
http://dx.doi.org/10.1128/AEM.02885-05
PMid:16820472 PMCid:PMC1489375
Wang, G. and Doyle, M. P. (1998). Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J. Food Prot., 61:662–667.
http://dx.doi.org/10.1016/S0014-5793(98)01090-4
http://dx.doi.org/10.1016/S0014-5793(98)00402-5
http://dx.doi.org/10.1016/S0014-5793(98)00423-2
http://dx.doi.org/10.1016/S0014-5793(98)00957-0
http://dx.doi.org/10.1016/S0014-5793(98)00272-5
http://dx.doi.org/10.1016/S0014-5793(98)01518-X
Wani SA, Pandit F, Samanta I, Bhat MA, Buchh AS (2004). Molecular epidemiology of shiga toxin producing Escherichia coli in India. Curr Sci 87: 1345–53.
WHO (1996) Generic protocol to estimate the burden of Shigella diarrhoea and dysenteric mortality. 1996; WHO/V&P/99.26. World Health Organization, Geneva, Switzerland.
Wright SK and Viola RE (2001). Alteration of the specificity of Malate Dehydrogenase chemical modulation of an active site Argenine. J Biol Chem 276: 31151–31155.
http://dx.doi.org/10.1074/jbc.M100892200
PMid:11389140
Yadav V, Joshi RK and Maurya SK (2009). Isolation and identification, Serotyping and antibiogram of Escherichia coli from diarrhoic poultry. J Vet Pub Hlth 07 : 145–147.