Advances in Animal and Veterinary Sciences
Research Article
Molecular and Conventional Detection of Antimicrobial Activity of Zinc Oxide Nanoparticles and Cinnamon Oil against Escherichia coli and Aspergillus flavus
Atef A. Hassan1 , Khairy F. Abo-Zaid2, Noha H. Oraby1*
1Department of Mycology and Mycotoxins; 2Serology Unit (Bacteriology), Animal Health Research Institute, Agriculture Research Center (ARC), Dokki, Cairo, Egypt.
Abstract | The antimicrobial activity of zinc oxide nanoparticles (ZnONPs) and cinnamon oils (C.O.) was evaluated by conventional and molecular methods against Aspergillus flavus (A.flavus) and Escherichia coli 0157 (E.coli) that recovered from cattle mastitis. In agar well diffusion method (WD), Minimum inhibitory concentration (MIC) of ZnONPs and C.O. for A.flavus was (100 μg/ml; 0.25%) and for E.coli 0157 were (50 μg/ml; 0.25%), respectively. The synergistic effects of these materials caused higher significant inhibition of all microbial growth by low and high doses by agar method. But, the molecular detection of virulent genes of E. coli (stx1) and A. flavus (AflR) by polymerase chain reaction (PCR) and the real-time PCR (RT-PCR) yielded uncorrelated results with WD tests. It is concluded that no direct correlation between WD, PCR, and RT-PCR and the WD tests are still inexpensive, eco-friendly, and rapidly applicable for screening of antimicrobials activity than genetic methods.
Keywords | Antimicrobial, Nanotechnology, Real-Time PCR.
Received | May 23, 2020; Accepted | June 03, 2020; Published | July 18, 2020
*Correspondence | Nora H Oraby, Department of Mycology and Mycotoxins; Cairo, Egypt; Email: [email protected]
Citation | Hassan AA, Abo-Zaid KF, Oraby NH (2020). Molecular and conventional detection of antimicrobial activity of zinc oxide nanoparticles and cinnamon oil against Escherichia coli and Aspergillus flavus. Adv. Anim. Vet. Sci. 8(8): 839-847.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.8.839.847
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Oraby et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Nowadays, the microbial infections resulted from bacteria and fungi are common and caused a significant reduction in the productivity of animals and the occurrence of human food poisoning (Quinn et al., 2002). Mastitis is the most common disease in dairy animals and resulted in significant economic losses due to a decrease in milk yield. Several studies recovered Staphylococcus aureus (S.aureus), E.coli, C. albicans, Aspergillus, and Penicillium species from mastitic milk of sheep and cattle as (Alyssa et al.,2012; Hassan et al., 2014). Hence, the development of more effective novel antimicrobial therapies is of great importance to prevent microbial infections particularly in dairy animals. Recently, the advances in nanotechnology enable synthesis metals nanomaterial which is used as an antimicrobial against common infectious agents without driving antibiotic resistance in organisms (Beyth et al., 2015; Mohan and Renjanadevi, 2016). The metals nanomaterial particularly ZnONPs have several applications as antimicrobial potentials against several pathogens (Liu et al., 2009 ; Hassan et al., 2015a). While, feed supplement with ZnONPs improves the immunity in dairy cattle (Sahoo et al., 2014) and have antimicrobial activity (Gong et al., 2007). These activities occur due to the penetration of nanoparticles into the cell membrane of organisms, generation of oxidative stress which destroys microbial cells (Seil and Webster, 2012). ZnONPs have significant effects as growth promotion, immune-modulatory, antibacterial and elevated efficiency of the reproduction in animals (Partha et al., 2015; 2016). However, the plant extracts and oils are nontoxic compounds and used in disease control replacing synthetic preservatives (Lee et al., 2007). The correlated detection of antimicrobials activities by agar diffusion tests and the molecular biology methods was previously evaluated (Chomvarin et al., 2004). They detected that disc diffusion (DD), microdilution tests (MD) were more applicable than genotyping RT-PCR. Therefore, this study was undertaken to evaluate the antimicrobial potentials of ZnONPs singly and/or in combination with cinnamon oil against recovered E. coli and A. flavus from dairy cattle mastitis. Moreover, the comparison between the conventional agar diffusion tests and molecular methods was investigated to evaluate the use of any of them in the rapid and effective detection of antimicrobial activities in large scales application.
Material and Methods
Samples
Two hundred samples (125 of mastitis milk and 25 of each of water, litter, and ration) were collected from dairy cattle farms in which animals suffered from mastitis. Milk and water samples were collected in sterile bottles, while, ration and litter samples taken in sterile polyethylene bags. All methods of collection and preparations were done as a method of (APHA, 2003). Each sample was divided into two parts and subjected for mycological and bacteriological examination.
Zinc Oxide Nanoparticles and Cinnamon Oil
ZnONPs were synthesized and characterized by the laboratory of ALDRIK Sigma chemical company, USA and it was in powder form with 50 nm particle size. While cinnamon oil was purchased in crud form from Al Gomhorya chemical company, Egypt.
Mycological Examination
Ten grams of finely grind rations and litter samples and 10 ml of milk sediment and water samples were added separately to 90 ml of 1% peptone water and stirred vigorously by electric blender for preparation of homogenate (APHA, 2003). One milliliter of homogenate was inoculated into Petri-dish plates and mixed with Sabouraud’s dextrose agar (SDA) and incubated 3-5 days at 25-28oC and identification of appeared mold and yeast colonies were identified according to, Pitt and Hocking (2009).
Bacteriological Examination
Samples were cultured onto MacConkey agar medium for 24 hrs at 37oC, then a peptone water cultures were prepared from appeared colonies to inoculate biochemical tests (Quinn et al., 2002). While, serological identification for E. coli species was undertaken according to (Neville and Bryant, 1986).
Antimicrobial potential OF ZnONPs and C.O. (Jin et al., 2009 and Jeff-Agboola et al., 2012).
The A.flavus and E.coli O157 that recovered from the present samples and the standard control of each were cultivated on (SDA and MacConkey agar) and incubated for (1-3 days at 28°C or 24 hours at 37°C), respectively. The spore suspension was prepared and counted in the hemocytometer slide. One ml of 105 spores of microbes was aseptically added to plates and covered with SDA medium (for fungus) and nutrient agar (for bacteria). Wells of 5 mm in Φ were made on surface of plates and add 100 µl of ZnO NPs (0, 25, 50, 100, 150, 200, 250 µg/ ml) or 100 µl of C.O (0, 0.25%, 0.5% , 1%, 2%, 3%) and incubated for 1-5 days at 28-37 oC.
Synergistic antimicrobial potential of znonps with C.O. :
In a separate 4 wells in plates, we added 50 µl of ZnONPs+ 50 µl of C.O. of the following concentrations: (0.25% of C.O.+ 100 µg/ml ZnONPs), (1% C.O+ 100 µg/ml ZnONPs), ( 0.25% C.O.+ 200 µg/ml ZnONPs) and (1% C.O.+ 200 µg/ml ZnONPs). Incubation of plates for 1-5 days at 28-37 oC. Then the plates were tested for the growth inhibitory zones around wells. All procedures were repeated 3 times to pooled data.
Detection virulent genes of E. coli O157 and A.flavus BY PCR. Preparation of treated strains of A.flavus and E. coli O157:
The A.flavus and E. coli O157 that recovered from the present samples were subjected to PCR detection of virulent gene expression before and after treatments with ZnONPs and C.O. In 50 ml sterile test tubs, add 20 ml of sterilized SD broth medium (for fungus) and nutrient broth (for bacteria) and 0.2 ml of 105 spore suspensions of 7 days old for all used microbes was inoculated into the tubes. Each strain was subjected for 6 doses treatments (low, 100 µg /ml ZnO NPs), (high, 500 µg/ml ZnONPs) (low, 0.25% C.O.), (high, 1%C.O.), (combination, 100 µg /ml ZnONPs+ 0.25% C.O.), (combination, 100 µg /ml ZnONPs+ 1% C.O.). The negative control was (Fusarium for A.flavus)( Staph aureus for E.coli O157) and the positive were (A,flavus and E.coli O157). All the tubes were incubated at 30°C for 3 days and kept at 5-8 °C till DNA extraction.
DNA extraction (Fittipaldi et al., 2012 and Hossain et al., 2015):
Genomic DNA of the strains was obtained using the genomic DNA Extraction Kit (Quick-DNA Miniprep DNA purification kit, cat. No. D3024) following the manufacturer’s instructions. DNA concentration was determined spectrophotometrically at 260/230 nm using SPECTROstar Nano” BMG LABTECH” and stored at −20°C until PCR amplification.
| Genes | Primer | Primer Design5-´-3´. | Amplicon bp |
| Stx1 |
stx1-F |
GACTTCTCGACTGCAAAGAC | 306 |
|
stx1-R |
TGTAACCGCTGTTGTACCTG | ||
|
AflR |
Afl R-F |
AACCGCATCCACAATCTCAT | 800 |
|
Afl R-R |
AGTGCAGTTCGCTCAGAACA |
Table 2: Standard cycling mode
| Step | Temperature | Duration | Cycles |
| UDG activation | 50°C | 2 minutes | Hold |
| Dual-Lock DNA polymerase | 95°C | 2 minutes | Hold |
| Denature | 95°C | 15 seconds |
40
|
| Anneal/extend | 60°C | 1 minute |
Table 3: Prevalence of fungal species i in samples of mastitis in cattle
|
Fungal Species |
Examined samples | ||||||||||
| Mastitis milk (125) | Letter (25) | Water (25) | Ration (25) | Total(200) | |||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | ||
| A. flavus | 20 | 16 | 9 | 36 | 19 | 76 | 8 | 32 | 56 | 28 | |
| A. niger | 10 | 8 | 2 | 8 | 4 | 16 | 4 | 16 | 20 | 10 | |
| A.ochraceous | 5 | 4 | 1 | 4 | 0 | 0 | 0 | 0 |
6 |
3 | |
| A.fumigatus | 10 | 8 | 2 | 8 | 0 | 0 | 0 | 0 | 12 | 6 | |
| Pencillium sp. | 25 | 20 | 3 | 12 | 0 | 0 | 5 | 20 | 33 | 15.5 | |
| Cladosporium sp. | 5 | 4 | 1 | 4 | 1 | 4 | 0 | 0 | 14 | 7 | |
| Mucor sp. | 0 | 0 | 3 | 12 | 0 | 0 | 3 | 12 | 6 | 6 | |
| Fusarium sp. | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 12 | 3 | 1.5 | |
| C. albicans | 20 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 10 | |
| Rhodotrella sp. | 15 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 7.5 | |
Tables 4: The incidence of bacterial species in samples of mastitis in cattle
| Bacterial Species | Type of examined samples | |||||||||
| Mastitic milk (125) | Letter (25) | Water (25) | Ration (25) | Total(200) | ||||||
| % | No. | % | No. | % | No. | % | No. | % | No. | |
|
E. coli (total) |
16 | 20 | 28 | 7 | 12 | 3 | 4 | 1 | 15 .5 | 31 |
|
E.coli O157 |
12 | 15 | 4 | 1 | 4 | 1 | 4 | 1 | 9 | 18 |
| E.coli O55 | 4 | 5 | - | - | - | - | - | - | 2.5 | 5 |
| E.coli O111 | 8 | 10 | 8 | 2 | 4 | 1 | 4 | 1 | 7 | 14 |
| E.coli O26 | 4 | 5 | - | - | 4 | 1 | - | - | 3 | 6 |
| C. freundi | - | - | 4 | 1 | 4 | 1 | - | - | 1 | 2 |
| C.diversus | - | - | 4 | 1 | - | - | - | - | 0.5 | 1 |
| K.pneumonia | 4 | 5 | 8 | 2 | 4 | 1 | - | - | 2 | 4 |
| K. oxytoca | - | 8 | 2 | - | - | - | - | 1 | 2 | |
| Ps.aeurginosa | 4 | 5 | 20 | 5 | 12 | 3 | 4 | 1 | 7 | 14 |
|
S. aureus |
8 | 10 | 12 | 3 | 8 | 2 | - | - | 7 | 7 |
PCR amplification (Somashekar et al., 2004 and Hossain et al., 2015):
The PCR amplification used primers for the detection of aflatoxin regulatory gene (aflR) of A.flavus and Shiga toxin gene (stx1) of E.coli O157 were prepared by Invitrogen Company (Table 1). The amplification conditions for AflR
gene were: 5 min initial step at 95 °C followed by 35 cycles at 95 °C for 30 sec, 56 °C for 30 sec and 72 °C for 30 sec and a final extension step at 72 °C for 10 min. While, the PCR amplification of Stx1 gene were: initial step at 95°C for 5 min, 25 cycles for 5s at 96 °C, 10 s at 54 ° C, 15 sec. at 68°C. Amplification products were electrophoresed with 1 ul of ethidium bromide per agarose gel added for visualization under UV light (1.5% w/v) (Sigma, USA), Using 100 bp DNA Ladder H3 RTU (Ready-to-Use) Cat. No. DM003-R500 from Gene Direx, Inc. Company, Litwania.
Real-Time PCR (Sharma and Nystromi, 2003 and Cruz and Buttner, 2008):
The RT-PCR was used to detect the DNA cycle threshold for aflR and stx1 genes using specific oligonucleotide primers and syber green Mix. The quantities were determined by RT-PCR in 20 µL containing 1 µL of DNA template, 10 µL of syber Green (Biosystems, and Catalog number: A25741). 7 µL PCR grade water, 1 µL of primer with a level of 10 pmol/µL. cycle was done in real-time PCR machine (Chrom4-BIO-RAD, USA), amplification condition as Standard cycling mode (Table 2)
Statistical Analysis
The obtained data were computerized and analyzed for calculation mean ± standard error according to SPSS 14 (2006).
Results and Discussion
In the current study, the most common recovered fungi was A. flavus from mastitis milk, letter, water and ration (16%, 36%, 76% and 32%), respectively. While, the yeasts spp. were detected only in mastitis milk samples as C. albicans and Rhodotorela sp. (16%,12%), respectively (Table 3). Whereas other genera of molds were recovered in variable frequencies. Aspergillus flavus constitute a public health hazard due to the production of aflatoxins which cause some degree of acute toxicity and are potential carcinogens (FDA,2000). On the other hand, E. coli potentiated the occurrence of bovine clinical mastitis (Hogan and Smith,2003) and recovered from milk and its product (Quinn et al., 2002). Herein, E. coli was the most predominant isolates from mastitis milk, letters, water, and animals’ ration (16%, 28%, 12%, and 4%), respectively. Currently, the strains of E.coli O157 and E.coli O111 were recovered from all examined samples at the rates of (12%, 8%) in mastitic milk, (4%, 4%) in water, (4%, 4%) in the ration and (4%, 8%) in letter samples respectively (Table 4). Seyffert et al. (2012) recovered S. aureus and E. coli from mastitic milk. Moreover, Parul et al. (2014) recovered E. coli from dairy animal litters, water and ration samples (8.33%, 3, 0%) and (3%, 11.11%, 0%) by Vanitha et al. (2018), respectively. The significant disease of E.coli O157: H7 which produced Shiga toxins (stx1 and stx2) is the hazard of food poisoning (Vali et al., 2007) and dairy cattle is the primary reservoir of infection (Perera et al., 2015). Hence, the discovery of the novel effective antimicrobial agents is required to overcome the microbial infections and resistance to commercial antibiotics (Whitesides, 2003). Today, ZnONPs have significant antimicrobial potentials and friendly safe to the environment (Violeta et al., 2011). Herein, A.flavus and E.coli O157 were the most prevalent isolates in samples of mastitis cattle and were subjected for investigation of the antimicrobial potentials of ZnONPs and C.O. by WD test. The (MIC) of ZnO NPs against A.flavus and E.coli O157 were (100, 50 μg /ml) and inhibition zones were (10±1.0, 13±0.7 mm), respectively (Table 5). In A.flavus, when ZnONPs levels increased from (100-900 µg/ml), the zones of inhibition also increased (10±1.0-30±3.0 mm).While, in E.coli O157, the inhibition zones elevated (13±0.7 to 29±2.0 mm), as levels of ZnONPs increased from (50- 900 µg/ml). The antimicrobial potential of ZnO NPs was detected against bacteria and fungi (Raghupathi et al., 2011; Hassan et al., 2017) that caused skin infection in buffaloes (Hassan et al., 2015b) and mastitis in cattle (E.coli and A. flavus) (Sabir et al., 2014). This is due to the penetration of ZnO-NPs the microbial cell wall, destruction and death of cells (Brayner, et al., 2006). Currently, MIC of C.O. against A.flavus and E.coli O157 were (0.25% for each) and inhibition zones increased as concentration levels increased (Table 6). El-Baroty et al. (2010) detected the significant antimicrobial activity of C.O. and the MIC values were ranged from (20-120 μg/ml). This activity due to C.O. rich with eugenol and cinnamaldehyde which enable them to penetrate the bacterial or fungal cell membrane and mitochondria cause cell death (Anwer et al, 2009) and impairment of cell enzyme system and cause gene toxicity (Abd El-Baky and El-Baroty, 2008). Recently, the awareness about the toxicity of nanoparticle applications resulted in significant attention for its conjugation with natural materials to avoid the toxic doses in animals. Currently, the synergistic effects of ZnONPs (100, 200 μg/ml) with C.O. (0.25% and 1%), resulted in significant growth inhibition of A.flavus and E.coli O157 (Table 7). The combination of low level of ZnONPs (100 μg /ml) with (0.25% C.O) caused increase in inhibitory zone (10±1.0 to15±2.0) and (17±0.2 to 20±0.8), respectively. While, elevation of C.O. concentration to (1%) at a low level of ZnONPs, increased significantly the inhibitory zones to (20±1.5 to 25±2.0), respectively. These results increase the availability of the application of nanomaterial in biomedicine by decreasing the used doses via conjuga
Table 5: Antimicrobial activity of Zinc oxide nanoparticles against A.flavus and E.coli 0157 recovered from cattle mastitis
| Examind isolates | Zones of inhibition (mm) at different concentration of Zinc oxide NPs (µg/ml) | |||||||||
| 50 | 100 | 200 | 300 | 400 | 500 | 600 | 700 | 800 | 900 | |
| A.flavus | ND | 10±1.0 | 15±0.5 | 15±1.0 | 17±1.5 | 20±0.8 | 22±2.0 | 27±2.5 | 30±1.0 | 30±3.0 |
| E.coli 0157 | 13±0.7 | 17±0.2 | 20±1.5 | 20±0.8 | 24±1.2 | 24±2.5 | 25±2.0 | 25±1.0 | 27±0.8 |
29±2.0 |
Table 6: Antimicrobial activity of cinnamon oil against A.flavus and E.coli 0157 recovered from cattle mastitis
| Tested isolates | Diameters of inhibitory zones( mm)/ gradual concentrations of cinnamon oil ( %) | |||||
| 0% | 0.25% | 0.5% | 1.0% | 2% | 3% | |
| A.flavus | ND | 5±0.5 | 10±1.5 | 11±0.5 | 15±0.8 | 18±1.0 |
| E.coli O157 | ND | 7±1.5 | 12±2.5 | 15±1.0 | 17±2.0 | 20±2.5 |
Table 7: Synergistic Antimicrobial activity of Cinnamon oil with ZnONPs against A.flavus and E.coli 0157 recovered from cattle mastitis
| Examined isolates | Diameters of inhibitory zones(mm) / combination of cinnamon oil and ZnONPs. | |||
|
100 μg/ml of ZnO NPs+ 0.25% of cinnamon oil. |
100 μg/ml of ZnO NPs+ 1.0% of cinnamon oil. |
200 μg/ml of ZnO NPs+ 0.25% of cinnamon oil. |
200 μg/ml of ZnO NPs + 1.0% of cinnamon oil. |
|
| A.flavus | 15±2.0 | 20±1.5 | 27±3.0 | 35±3.0 |
| E.coli O157 | 20±0.8 | 25±2.0 | 25±2.5 | 35±2,0 |
Table 8: Comparison between agar diffusion tests, PCR amplification and RT-PCR for detection AflaR -gene of A.flavus and stx1-gene of E.coli O157 that treated with ZnONPs singly or in combination with C.O.
|
Treatments Trials |
Agar Diffusion Test |
AflaR -gene of A.flavus |
stx1-gene of E.coli O157 |
||||||
|
PCR |
|
C.T. (RT-PCR) |
PCR
|
C.T. (RT-PCR) |
|||||
| L |
H |
L |
H |
L | H | L |
H |
||
|
Control (Untreated) |
G |
P |
|
26.33 |
P |
|
19.46 |
||
|
ZnONPs |
NG |
N |
N |
26.62 |
23.26 |
P |
P |
24.01 |
23.99 |
|
C.O. |
NG |
P |
P |
23.41 |
22.57 |
N |
P |
14.72 |
20.01 |
|
Combination of C.O.+ ZnONPs |
NG |
N |
P |
28.34 |
28.39 |
P |
P |
20.43 |
24.07 |
- G: growth of microbial cells -NG: no growth -P: positive DNA band -N: no DNA band - C.T.: Cycle Threshold of DNA - ZnO NPs: 100 µg/ml (Low dose), 500 µg/ml (High dose). - C.O.: 0.25% (low dose), 1% (high dose). -.Combination : 100 µg/ml of ZnONPs+ 1% C.O( high dose), 100 µg/ml of ZnONPs+ 0.25% C.O. (low dose).
tion with natural materials. Similarly, Hassan et al. (2019) detected the MIC of ZnONPs against Fusarium sp. was (500 μg/ml) and significantly decreased to (100 μg /ml) when combined with curcumin or probiotic (0.25% for each). This enables to prevent drug resistance and resulted in significant antimicrobial efficacy (Chow and Yu, 1999).
In the present study, PCR detection of the virulent genes in isolated A. flavus (aflR) and E. coli O157( stx1) from cattle mastitis (4 representative isolates). The DNA bands of 2 isolates of A.flavus were similar to the standard strain, while others showed no bands for aflR gene (Figure 1). The expression of the stx1 gene of E.coli O157, 2 isolates not showed any DNA fragment and other isolates were positive for the stx1 gene similar to the standard strain (Figure 2). Cruz and Buttner (2008) detected the alfR gene in A.flavus by PCR and different results of DNA bands occurred. While, Scherm et al. (2005), detected alfR and alfQ in A.flavus isolated from animal feeds. PCR detection of virulent genes stx1 and stx2 in E. coli isolated food and beef samples (Godambe et al., 2017) that cause food-born infection (Ferens and Hovde, 2011). Currently, the positive isolates of virulent genes were used for PCR detection of antimicrobial potentials of ZnONPs and C.O. (Figures, 1, 2). The PCR amplification of DNA bands of control for each used isolate was similar to the general characters of a standard reference untreated species of A.flavus and E.coli O157 (Figures 3, 4). Whereas, treating A.flavus by low (100 g/ml) and high (500 g/ml) doses of ZnO NPs eliminated the signals of DNA bands (Figure 3). But DNA bands were observed in the case of E. coli O157 with low or high doses of ZnO NPs (Figure 4). The C.O. effects on genes of A.flavus, either at low and high doses (0.25%, 1%), not cause any changes in DNA bands signals. On the contrary, the treatment of E.coli O157 with a high dose of C.O.(1%) resulted in the absence of DNA band, but a low dose (0.25%) not cause any changes. Whereas, the combination of ZnO NPs and C.O. presence of DNA band in E. coli. While, there was low faint DNA band in treatment of A.flavus with (100 μg /ml of ZnONPs+ 1% C.O.). Recently, the RT-PCR help in the generation of a specific fluorescent signal in real-time analysis and quantitation of DNA targets (Schena et al., 2004) and allow rapid, sensitive, specific, and high accurate activity than traditional DNA-PCR method (Bilodeau, 2011). Herein, the RT-PCR system directed against DNA extracted from isolates of A.flavus and E.coli 0157 was done (Figures 1-4 and Table 8). The treatment doses of ZnONPs alone or in combination with C.O. increased the DNA cycle threshold (C.T). The treatments of A.flavus with ZnO NPs (100g/ml) resulted in a significant increase in DNA C.T. values (26.62, 28.34) higher than that DNA of non-treated isolates (26.33).
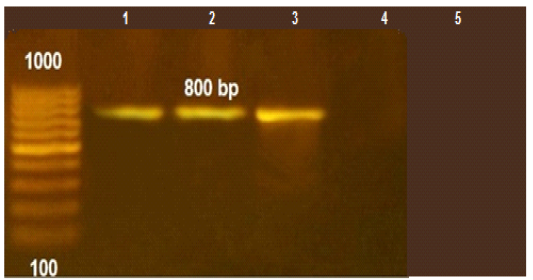
Figure 1: PCR amplification for aflaR gene of A.flavus(at 800pb) Lane L:100 bp DNA ladder standard. Lane 1: Positive control of A.flavus. Lane 2-5 A.flavus isolated from mastitis

Figure 2: The PCR amplification for stx1gene of E.coli O175(at 306pb).Lane R: 100bp DNA ladder standard; Lane 1: Positive control of E.coli 0157., Lane 2-5: the isolates from mastitis
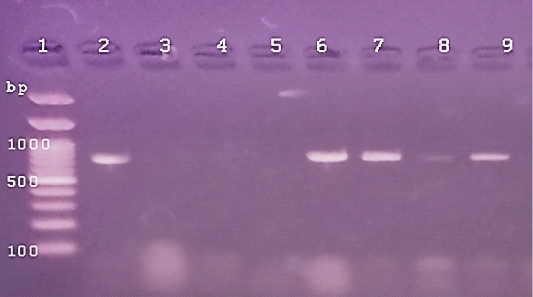
Figure 3: The PCR amplification for aflaR gene of A.flavus(at 800pb) Lane 1:100 bp DNA ladder standard. Lane 2: Positive control of A.flavus.Lane 3: Negative control (Fusarium sp.) Lane 4: Treated 100 μg /ml of ZnONPs).Lane5: Treated 500 μg /ml of ZnONPs). Lane 6: Treated 0.25% C.O. Lane 7: Treated 1% C.O. Lane 8: Combination treat. of 100 μg /ml of ZnONPs+1%C.O. Lane9: Combination treat. of 100 μg /ml of ZnONPs+ 0.25% C.O.
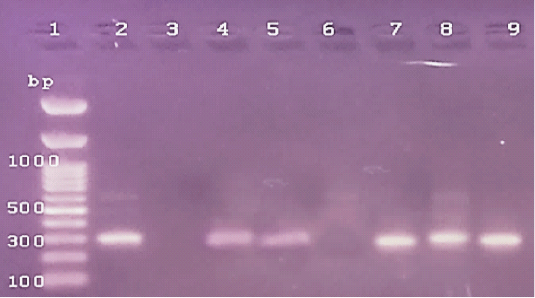
Figure 4: The PCR amplification for stx1gene of E.coli O175(at 306pb).Lane1: 100bp DNA ladder standard; Lane 2: Positive control of E.coli 0157.Lane 3: Negative control (S.aureas) Lane 4: Treated 100 μg /ml of ZnONPs). Lane 5: Treated 500 μg /ml of ZnONPs). Lane 6: Treated 0.25% C.O. Lane 7: Treated 1% C.O.Lane 8: Combination treat. of 100 μg /ml of ZnONPs+ 1% C.O. Lane 9: Combination treat. of 100 μg /ml of ZnONPs+ 0.25% C.O.
While, the treatment of E.coli with low and high doses of ZnONPs caused a treatment with C.O. caused a decrease in DNA C.T. values (Table 8). It is suggested that the higher DNA C.T. due to the lower number of DNA copies of the genes in treated isolates with ZnONPs than that of untreated ones and contrary to this were reported in C.O. (Table 8). Several studies used RT-PCR for rapid detection of genes pathogens as Sharma and Nystromi (2003) and Hu et al. (2020) for stx1 and stx2 in E.coli O157: H7 in food, Scherm et al. (2005), for detecting aflatoxin regulatory genes.Copping et al. (2005) found that the inhibitory concentration of antifungals against C.albicans elevated the activity of secreted proteinase (Sap) and SAP genes detection by RT– PCR. Labeed et al. (2016) and Hassan et al. (2017) detected the absence of DNA band in PCR of Afla gene after bio-control of mycotoxigenic A. flavus and no levels observed in chemical detection of AFB1. Rathore et al. (2018) found that variety in the correlation between disc diffusion and genotypic PCR antibiotic sensitivity pattern against virulent genes in S. aureus that ranged from (58.3%-100%). While, Zheng et al. (2015) resulted in significant antimicrobial activity of Enterococcus faecium against gram-positive and gram-negative bacteria using agar well diffusion method as a simple method and PCR amplification not detected any tested virulence genes. Furthermore, Chomvarin et al. (2004) detected that disc diffusion and agglutination tests are of the highest sensitivity and specificity and both assays are technically simple and can be easier to perform in routine laboratories than PCR in the evaluation of drugs against bacterial and fungal pathogens. Hence, the evaluation of antimicrobials by covenantal method as agar WD yielded significant specific results and despite advances in PCR and RT-PCR we detected variable in accord findings.
Conclusion
From the forgoing results it is concluded that the antimicrobial potential of ZnONPs was more effective than traditional antibacterials as oil and resulted in decreased and eliminated the targeted DNA gene expression of aflatoxigenic A.flavus and E.coli. In addition the RT-PCR confirmed these changes by increase in DNA cycle threshold . The synergistic action of ZnONPs with natural oil caused significant antibacterial potential and resulted in decrease the used doses of nanomaterial , hence, we can overcome nanomaterial toxicity for future application in veterinary medicine. The conventional laboratory diffusion tests are still most satisfactory, simple and inexpensive in comparison with genotyping methods as PCR and RT-PCR. Hence, nanotechnology has huge significant progressive advancement in biotechnology and biomedicine related to human and animal science as increase the safety of their health, production and hence elevation of national income.
Conflict of interest
All authors declare that there no conflict of interest.
Acknowledgment
The authors are gratefully acknowledged to Prof. Dr. Hazem Hassan Mahmoud, for his kind assistance and fund in identification and characterization of the prepared and used zinc nanoparticles.
authors contribution
Atef Hassan conceived and designed the experiments; Noha Oraby and K.Abo-zaid did the methodology, Atef Hassan and Noha Oraby did manuscript preparation, analyzed the data and wrote this manuscript; Corresponding author N.Oraby. All authors read and approved the final manuscript.
References
14.0.0, and 12 June, 2006. Standard Version, Copyright ® SPSS Inc. USA.






