Advances in Animal and Veterinary Sciences
Research Article
Genetic Diversity among Different Samples of Camel’s Tick (Hyalomma dromedarii) in Taif city, Saudi Arabia
Bander Albogami*
Department of Biology, College of Sciences, Taif University, 21974 Taif, P.O. Box 888, Saudi Arabia.
Abstract | The ticks (Hyalomma dromedarii) are significant disease transmit vectors to camel in Saudi Arabia. They transfer a range of pathogens such as bacteria, protozoa and viruses to people and animals. Their significance as a malady vector requires dependable molecular technique in the present research, the genetic variability of H. dromedarii from six farms located in Taif governorate of Saudi Arabia were investigated by using ISSR marker technique. Eight ISSR markers were chosen to deliver clear and reproducible polymorphic loci. The genetic variability within and among all samples of H. dromedarii were investigated via these markers. A total of 102 DNA loci were produced. Out of these loci, 63 were with ratio of 61.8%. The number of total bands varied from 8 to 16 bands, and the bands size ranging from 120 to 2100 bp. By using Neighbor-joining genetic distance method, the 12 samples were assembled into two main group with 0.56 genetic similarity. The first group contains two samples, number 2 and 11, from different farm. The other ten samples are located in second group which divided into two clusters. The value of genetic similarity varied from 0.877 for two samples, tick-9 and tick-12 to a lowest value of 0.056 for samples tick-3 and tick-11. The present data indicates that there is no remarkable correlation between the geographic distance and genetic similarity.
Keywords | Genetic diversity, Hyalomma dromedarii, ISSR-PCR, Taif city, Saudi Arabia
Received | November 07, 2019; Accepted | February 17, 2020; Published | March 03, 2020
*Correspondence | Bander Albogami, Department of Biology, College of Sciences, Taif University, 21974 Taif, P.O. Box 888, Saudi Arabia; Email: [email protected]
Citation | Albogami B (2020). Genetic diversity among different samples of camel’s tick (Hyalomma dromedarii) in Taif city, Saudi Arabia. Adv. Anim. Vet. Sci. 8(3): 285-289.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.3.285.289
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Albogami. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Hyalomma dromedarii is a common camel tick that distributes in deserts, plains and semi-plains from the southern of Soviet Union and northwest India to Saudi Arabia and North Africa to the equator wherever camels are found (Hoogstraal et al., 1981). Temperature and relative humidity are the most important factors affecting the tick’s life cycle and growth period (King et al., 1988; Pegram and Banda, 1990). Temperature and relative humidity also affect egg production, hatchery and crushing ratios (El-Ghali et al., 2003). The life cycle of ticks has been studied in laboratory conditions only in order to avoid unfavorable environmental conditions, multiple growth patterns and multiple host (Hagras and Khalil, 1988; Alahmed and Kheir, 2003). The development of H. dromedarii in natural conditions was also studied in order to determine the finest times of tick development (El-Ghali and Hassan, 2009). It has been reported that H. dromedarii is able to move between three hosts, changing to 2 hosts under heat pressure to avoid drying of larvae (Alahmed and Kheir, 2003). Also, 60% of H. dromedarii changed to two-host when fed on rabbits (El-Ghali and Hassan, 2010). The Hard tick (Acari: Ixodidae) is among the group of arthropods and vectors of many pathogenic microorganisms such (Borrelia, Anaplasma, Ehrlichia, Babesia, Theileria and Hyalomma), which affect both (humans and animals) (El-Ghali and Hassan, 2009). Ticks are a major factor limiting animal production, and thus diseases are transmitted to cattle (Estrada-Peña and Jongejan, 1999; Parola and Raoult, 2001). Ticks cause many diseases such as paralysis, toxins, irritability, allergies and a wide variety of infectious diseases (El-Ghali and Hassan, 2009; Parola and Raoult, 2001; Elston, 2010; Briciu et al., 2011; Roderick, 1996). In Saudi Arabia, camel tick (H. dromedarii) affected negatively on the health and productivity of camels. As a result of tick infection at high rates gives the tick economic importance (Diab et al., 2001; El-Ghali and Hassan, 2009).
Molecular methods have recently revolutionized systematic insects, which are progressively useful for bugs and ticks. The use of molecular markers has turned out to be increasingly significant in the systematic investigation of ticks (Liu, 2012; Léger et al., 2015). The principles and characteristics of several molecular markers such DALP, RAPD (Edwardsa et al., 1997), RFLP (Osakabe and Sakagami, 1994), AFLP (Zhang et al., 2008), SSR (Navajas et al., 2002) and sequencing technique (Cruickshank, 2002) have been widely applied in the molecular study of mites. ISSR is one of the most molecular markers technique that used in the systematic study of ticks (Zou et al., 2011). In this technique, single initiators scattered sequences along the genome are used, which is cheaper, faster and easier to conduct than some other markers (Zou et al., 2011). These markers give a new kind of different unique fragments that differentiate individuals at the DNA sequence level, most of which are polymorphic (Nagaoka and Ogihara, 1997). ISSR is dominant markers of heredity and can produce huge quantities of instructive and reproducible alleles. Until this point in time, there are no reports accessible to utilize ISSR markers to study H. dromedarii in camel. Therefore, the targets of this investigation are mainly to test the effectiveness of ISSR markers as a molecular technique analysis to check the genetic diversity within and between tick populations.
Material and Methods
Ticks collection and DNA isolation
Male and female adult ticks (H. dromedarii) of a total 12 individual samples were collected from three different regions of Taif city, four from each farm. The three places were close to each other by about 5 km in distance between each other and located in the central of Taif city. Every tick sample was washed in 70% ethanol once with distilled water and then left to dry before DNA isolation. Individual single tick sample was used for genomic DNA isolation using DNA easy Mini Kit (QIAGEN, Germantown, MD 20874, USA) according to the instructions of the manufacturer. The DNA samples were frozen until use.
ISSR–PCR amplification
For ISSR analysis, eight primers were used. All primers information is outlined in Table 1. PCR amplification of ISSR was carried out according to Hassan et al. (2014), in 25 μl volume using Go Taq® Green Master Mix, Promega, USA. DNA amplification carried out using the C1000TM Thermo Cycler Bio-Rad, Germany. Primer annealing temperature was 50° C for 1.5 min. The analysis of the resulted DNA fragments was visualized using 1.5% agarose gel. DNA was photographed by a Bio-Rad Gel Doc 2000 device.
Table 1: ISSR primers names and sequences used in the present study.
|
Serial number |
Primers name |
Primers sequences 5' → 3' |
| 1 | ISSR-2 | GAG AGA GAG AGA GAG AA |
| 2 | ISSR-4 | GAG AGA GAG AGA GAG ATT |
| 3 | ISSR-9 | TAG CAC ACA CAC ACA CA |
| 4 | ISSR-10 | CAG CAC ACA CAC ACA CA |
| 5 | ISSR-14 | GAG AGA GAG AGA GAG AC |
| 6 | ISSR-16 | GAG AGA GAG AGAG AGA C |
| ISSR-19 | AGA GAG AGA GAG AGA GTT | |
| 8 | ISSR-20 | GAGAGAGAGAGAGAGAAT |
Data analysis
The ISSR product were analyzed using NTSYS-PC program version 2.01 (Rohlf, 2000). The presence loci were recorded for “1” and the absence loci as “0”, while missing data take “9”. The genetic relationship between the samples was measured by calculating the Jaccard’s similarity coefficient dependent on the number of common loci ranges that amplified by the primers. The alteration matrix was formed using the neighbor-joining method, thus the dendrogram was reconstructed.
RESULTS
The electrophoresis banding pattern showed that the eight markers of ISSR could demonstrate high level of diversity existing among the genotypes consequently; the markers were functional for each of twelve genotypes of the ticks (H. dromedarii) (Figures 1 and 2). PCR-based molecular markers assume a significant role in the investigation of genetic variation in these species. The used primers of ISSR generated 102 bands ranging from 120 to 2100 bp (Table 2 and Figure 1), which were polymorphisms up to 61.8 % and lower level of monomorphic with 38.2%. The maximum and minimum number of amplified bands ranged from 16 and 8 that belonged to ISSR-19 and ISSR-8, respectively. The polymorphism value was varied from 12.5 % to 87.8 %. The average band per primer was 12.75 and the average percentage of monomorphic bands (38.2 %). As indicated by genetic similarity and intraspecific differentiation, the 12 individuals were gathered into two main groups with about 0.56 genetic similarity (Figure 2). Interestingly, the first main group contained sample number 2 (from farm number 1) and 11 (from farm number 3) only. The second group was contained two clusters, the first one contain sample number 3 only, while the second one having three sub-cluster that contained samples numbers 1, 4, 5, 6, 7, 8, 9,10, and 12 (Figure 2). The utilization of molecular markers was planning to illustrate quick and solid segregation of genetic relations of H. dromedarii. In the present study, DNA fingerprinting data generated by using ISSR markers were found to be efficient to distinguish different tick individual. In this case, a set of minimum number of primers are required to be selected for the varietal identification and to select a set with minimum number of primers, it is important to consider level of marker polymorphism. In case of ISSR markers, 61.8% genetic polymorphism was observed, showing that ISSR markers have high potential compared to RAPD marker in discriminating H. dromedarii types.
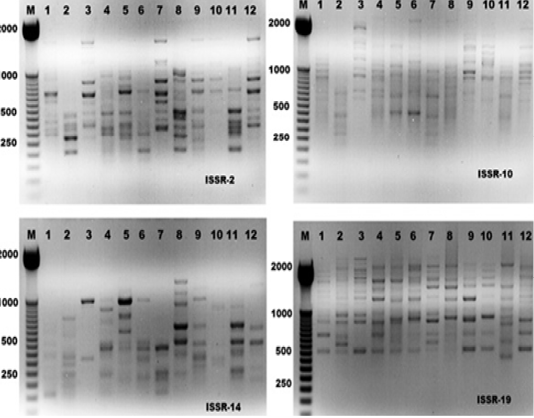
Figure 1: ISSR-PCR profiles of the twelve Hyalomma dromedarii samples that generated with four ISSR primers; ISSR-2, ISSR-10, ISSR-14 and ISSR-19, respectively. First lane on each panel is 50 bp molecular weight markers.
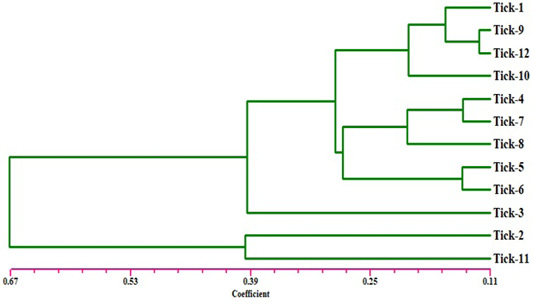
Figure 2: Dendrogram analysis showing relationship of the twelve samples of Hyalomma dromedarii based on ISSR marker using Jaccard’s similarity coefficient. Individuals from 1 to 4 are from farm number 1 and individuals 5 to 8 from farm number 2, while individuals 9 to 12 from farm number 3.
Eight selected ISSR markers revealed the considerable intraspecific variability among 12 individuals of H. dromedarii (Table 3). The values of genetic similarity depend on ISSR markers technique were gotten by multivariable investigation using Nei’s coefficient (data not shown). The results of genetic similarity from all twelve samples of H. dromedarii were ranged from 0.944 to 0.123 (Table 3). The genetic diversity index of sample number 2 and 11 represented the highest one within total individuals. As shown in Figure 2, the dendrogram demonstrating genetic relationship among H. dromedarii individuals based on the total number of amplified ISSR loci. The genetic variety index and genetic similarity rate of samples 3 and 11 were remarkably different (P<0.05) from each other. The average genetic diversity index with geographic origins was 0.123.
Table 2: Polymorphic bands of each ISSR primer and percentage of polymorphism between the twelve Hyalomma dromedarii samples.
| Primers | Total bands | No. of monomorphic bands | No. polymorphic bands | % Monomorphic bands | % Polymorphic bands |
| ISSI-2 | 15 | 5 | 10 | 33.3 | 66.7 |
| ISSI-4 | 11 | 2 | 9 | 18.2 | 81.8 |
| ISSI-9 | 12 | 10 | 2 | 83.3 | 16.7 |
| ISSI-10 | 14 | 2 | 12 | 14.3 | 85.7 |
| ISSI-14 | 14 | 3 | 11 | 37.5 | 62.5 |
| ISSI-16 | 12 | 2 | 10 | 16.7 | 83.3 |
| ISSI-19 | 16 | 14 | 2 | 87.8 | 12.5 |
| ISSI-20 | 8 | 1 | 7 | 12.5 | 87.8 |
| Total | 102 | 39 | 63 | 38.2 | 61.8 |
DISCUSSION
The present research was conducted to investigate the genetic diversity among different individuals of H. dromedarii that collected from three different farms using ISSR marker technique because of its great level of polymorphism marker at DNA level (Zietkiewicz et al., 1994). ISSR markers technique can be utilized in genetic diversity between samples of same species because they adequately identify extremely low degrees of genetic variety (Zietkiewicz et al., 1994). Clustering investigation by UPGMA confirmed that H. dromedarii samples of similar location could not be gathered together to produce same clade (Figure 2). As shown in Figure 2, individuals 1 through 4 who were collected from one farm fell into completely different clade. Therefore, the eight ISSR markers were successfully used to differentiate the individuals from same farm, indicating that ISSR markers are appropriate method for intraspecific recognition. As opposed to other PCR-based methods like RAPD, ISSR has the advantage of more nucleotides primer sequence that it allows for additional stringent annealing temperatures (Navajas and Fenton, 2000). These higher annealing temperatures clearly give a higher dependability
Table 3: Genetic similarity of the twelve Hyalomma dromedarii samples based on ISSR markers.
| Tick1 | Tick2 | Tick3 | Tick4 | Tick5 | Tick6 | Tick7 | Tick8 | Tick9 | Tick10 | Tick11 | Tick12 | |
| Tick1 | 0.000 | |||||||||||
| Tick2 | 0.877 | 0.000 | ||||||||||
| Tick3 | 0.403 | 0.522 | 0.000 | |||||||||
| Tick4 | 0.238 | 0.619 | 0.433 | 0.000 | ||||||||
| Tick5 | 0.342 | 0.844 | 0.371 | 0.2057 | 0.000 | |||||||
| Tick6 | 0.437 | 0.557 | 0.476 | 0.205 | 0.143 | 0.000 | ||||||
| Tick7 | 0.176 | 0.834 | 0.419 | 0.141 | 0.367 | 0.367 | 0.000 | |||||
| Tick8 | 0.423 | 0.607 | 0.644 | 0.166 | 0.311 | 0.237 | 0.247 | 0.000 | ||||
| Tick9 | 0.192 | 0.781 | 0.279 | 0.222 | 0.160 | 0.314 | 0.235 | 0.259 | 0.000 | |||
| Tick10 | 0.270 | 0.773 | 0.299 | 0.301 | 0.238 | 0.333 | 0.295 | 0.406 | 0.162 | 0.000 | ||
| Tick11 | 0.605 | 0.396 | 0.944 | 0.530 | 0.573 | 0.573 | 0.611 | 0.373 | 0.559 | 0.753 | 0.000 | |
| Tick12 | 0.133 | 0.877 | 0.220 | 0.238 | 0.255 | 0.342 | 0.176 | 0.423 | 0.123 | 0.183 | 0.605 | 0.000 |
and reproducibility results. Moreover, AFLP technique has several disadvantage points like several intensive laboratory steps and has a highly costs (Navajas and Fenton, 2000). On other hand, microsatellites technique like ISSR are more specific and produce more polymorphous loci (Coates et al., 2002). However, they need to know the DNA sequence to synthesis the exact sequence that are restricted mainly to economically species. Up-to-date, genetics studies recommended that the degree of genetic variety of a species is strictly linked with its adaptability, attainability and evolutionary possibility, and therefore genetic diversity is essential for organisms adjust to environmental change. Here, the genetic similarity value was ranged from 0.944 between individual 3 and 11 to 0.123 between individuals 9 and 12. These values designated a substantial reduction of genetic variety, most likely because of the high genetic isolation of the twelve samples that examined. Interestingly, individual number 11 showed high genetic diversity amongst all samples, probably due to random genetic drift.
Regardless of the constant distribution of H. dromedarii, the twelve individuals signify genetically differentiated indicating more genetic variations involved in all individuals from the three different farms. These data proposing that the existence of high genetic diversity among H. dromedarii individuals. The dendrogram that derived from using eight primers of ISSR showed that the ability of ISSR markers to group the twelve individuals into different clusters. These data were in agreement with several reports published previously with other organisms (Wang et al., 2005; Sica et al., 2005).
CONCLUSION
In conclusion, the loci that produced by the eight primers of ISSR permit to identify the genetic polymorphism among individuals of H. dromedarii, being helpful to decide the genetic diversity of this species. As a result, there was a high genetic variability among H. dromedarii individuals that collected from same or different locations.
Authors Contribution
Design and conduct the experiments, as well as writing, review, and editing the manuscript were done by Bander Albogami.
Conflict of interest
The author declared that there is no conflict of interest.
REFERENCES






