Advances in Animal and Veterinary Sciences
Research Article
Polymorphism of Candidate Genes Related to the Number of Teat, Vertebrae and Ribs in Pigs
Son Trinh Hong2, Vinh Nguyen Thi1, Pham Pham Duy2, Luc Do Duc1, Dang Pham Kim1, Giang Nguyen Thi Phuong1, Tuan Nguyen Ngoc Minh3, Thinh Nguyen Hoang1*
1Faculty of Animal Science, Vietnam National University of Agriculture, Vietnam; 2Thuy Phuong Pig Research and Development Center, Vietnam National Institute of Animal Science, Vietnam; 3Hung Vuong University, Vietnam.
Abstract | Prolactin receptor (PRLR) and vertnin (VRTN) gene is considered as genetic marker for teat, vertebrae and ribs number in pigs. In this study, the polymorphisms of these genes was investigated in Landrace, Yorkshire and Meishan purebred pigs. A total of 102 animals including 37 Landrace, 35 Yorkshire and 30 Meishan were collected from Thuy Phuong Pig Research and Development Center, Vietnam National Institute of Animal Science, Vietnam. Pig individuals were genotyped with PCR and PCR-RFLP methods. Allelic and genotypic frequencies were calculated. Hardy-Weinberg equilibrium (HWE) was performed using χ2-test. For PRLR, 3 genotypes (AA, AB and BB) were detected and the most abundant genotype was the heterozygous one. The frequencies of expected genotype AA were 0.35, 0.40, and 0.17 for Landrace, Yorkshire, and Meishan, respectively. Frequencies of allele A were 0.61 (Landrace), 0.69 (Yorkshire), and 0.59 (Meishan). The loci were in HWE for all group of pigs except Meishan. For VRTN, those values for QQ genotype were 0.30 (Landrace), 0.37 (Yorkshire), and 0.10 (Meishan), while Q allele frequencies were 0.43 (Landrace), 0.57 (Yorkshire), and 0.22 (Meishan). HWE was detected for Meishan Pig.
Keywords | Prolactin receptor, Vertnin gene, Teat, Vertebrae, Rib
Received | October 18, 2019; Accepted | February 17, 2020; Published | March 03, 2020
*Correspondence | Thinh Nguyen Hoang, Faculty of Animal Science, Vietnam National University of Agriculture, Trau Quy, Gia Lam, Ha noi, Vietnam; Email: [email protected]
Citation | Hong ST, Thi VN, Duy PP, Duc LD, Kim DP, Phuong GNT, Minh TNN, Hoang TN (2020). Polymorphism of candidate genes related to the number of teat, vertebrae and ribs in pigs. Adv. Anim. Vet. Sci. 8(3): 229-233.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.3.229.233
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Hong et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Improvement reproduction has become of great interest in pig industry as good fecundity is directly related to a sow’s productive life. An important trait related to the success of pig reproduction is the number of teats and vertebral because it reflects directly the mothering ability of sows (Brisbane and Chesnais, 1996; Hirooka et al., 2001; Stalder et al., 2000), which is a limiting factor for the increased number of weaned piglets. However, most reproduction traits including teat number are rather difficult to enhance through quantitative selection methods because of their low heritability and their sex-limited expression (Clayton et al., 1981; McKay and Rahnefeld, 1990). The applied of molecular genetics map in livestock production over the last 10 years has allowed detecting genomic regions contributing to the genetic variation of quantitative traits. These are known as quantitative trait loci (QTL). The use of genetic marker information may then be very useful to increase rates of genetic improvement. This is particularly true for reproductive traits, since simulation studies have demonstrated that the efficiency of marker-assisted selection relative to phenotypic selection is greatest for lowly heritable and sex-limited traits (Lande and Thompson, 1990).
Prolactin is a polypeptide hormone secreated by the anterior pituitary gland and has been shown to invole in many different endocrine activities and is essential for reproductive performance, mammary development and lactation. The hormone exerts its physiological effects via the prolactin receptor (PRLR) which has been detected in various tissues in many mammalian species (Kelly et al., 1991). The porcine PRLR gene has been mapped to chromosome 16 and Alu I polymorphism in the PRLR gene associated with litter size traits (Vincent et al., 1997; Rothschild et al., 1998; Southwood et al., 1999; Drogemuller et al., 2001; van Rens and van der Lende, 2002; Putnova et al., 2002; van Rens et al., 2003; Terman, 2005). Allele A has a significant effect on the number of piglets born alive (Drogemuller et al., 2001). Vertnin (VRTN) gene has been mapped to porcine chromosome 7 (Ren et al., 2012) and associated with the number of vertebrae and teat (Borchers et al., 2004; Wang et al., 2011; Yang et al., 2016). European commercial pigs as Large White, Landrace and Ducroc have more thoracic, lumbar vertebrae (Borchers et al., 2004; Wang et al., 2011; Yang et al., 2016) and teat number (Yang et al., 2016). Yang et al. (2016) indicated in the Chinese indigenous pigs, the QW pigs had more vertebrae (14.34 vs 13.91) and teats (20.81 vs. 20.31) than WW pigs; and in the Landrace pigs, the number of teats of QQ pigs (12.71) have more teats than QW pigs (12.35). Untill now, there are not many studies evaluating the relationship between PRLR and VRTN gene with teat vertebral and ribs number of pigs. The aim of this study is to initially investigate the polymorphism of PRLR and VRTN genes of three breed pigs in North Vietnam.
MATERIALS AND METHODS
The breeds (French Landrace and Yorkshire, and Danish Meishan) used in this study are came from Thuy Phuong Pig Research and Development Center, Vietnam National Institute of Animal Science, Vietnam. Ear tissue samples of 102 individuals pigs (37 Landrace, 35 Yorkshire and 30 Meishan) were collected and kept in a centrifuge tube (1,5ml), and stored at -20oC until DNA extraction. Genomic DNA was extracted using QIAamp DNA Tissue Kit. The PRLR gene fragment was amplified using PCR with designed primer reported by (Drogemuller et al., 2001). Particularly, the forward primer was 5’-CGT GGC TCC GTT TGA AGA ACC-3’and reverse primer was 5’-CTG AAA GGA GTG CAT AAA GCC-3’. The VRTN gene was amplified using primer publised by Yang et al. (2016). The forward and reverse was 5’-GGC AGG GAA GGT GTT TGT TA-3’ and 5’-GAC TGG CCT CTG TCC CTT G-3’, respectively.
The PCR reactions was performed using a reaction mix of 25µl containing 50ng of DNA genomic, 0.5µM primers of each, 0.2 mM dNTPs, 1.5mM MgCl2, 2.0 Taq polymerase and buffer. Amplification conditions of genes were 94oC for 4 min, followed by 35 cycles of 94oC for 45 sec, 50oC for 45 sec, 72oC for 1 min, with a final extension at 72oC for 10 min. The quality of PCR products were checked by 1% agarose gel electrophoresis.
For genotyping of PRLR, an AluI PCR-RFLP polymorphism was identified in porcine 163 bp fragment of the gene. 8.5μL of PCR product was digested at 37°C for 4 hours in a total volume of 30μL, containing 2 UI of the appropriate restriction enzyme, 3μL of restriction buffer, and 18.3μL of H2O and fragments were separated on agarose gel. For genotyping of VRTN, PCR products were separated by 2% agarose gel electrophoresis and the genotypes were visually recorded according to the length of amplicon.
The Hardy-Weinberg equilibrium in each breed was tested by comparing the expected and observed genotype count in using chi-squared test (χ²). Data were analyzed by using Proc Freq with SAS software,version 9.1. P- values <0.05 were considered as significant.
RESULTS
Genotyping
The results of PRLR/Alu I polymorphism detection by PCR-RFLP method are showed in Figure 1. Two alleles (A and B) and three genotypes (AA, AB and BB) were observed in three breeds. The length of restriction fragments were as follow: 85, 59, and 19 bp for allele A and 104 and 59 bp for allele B. This result was consistent with reported of Drogemuller et al. (2001), Omelka et al. (2008). For VRTN gene, the mutant allele (insert, Q allele) was represented by amplicons of 411bp and the wild type allele (delete, W allele) by amplicons of 120 bp. Consequently three genotypes QQ, QW and WW were found (Figure 2). These finding are similar those found in a previous study of Yang et al. (2016).
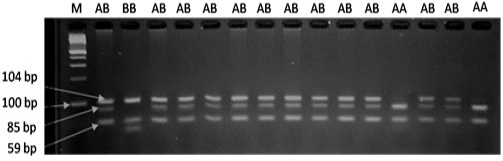
Figure 1: PCR fragments of PRLR gene digested by AluI on 3% agarose gel. The genotypes are shown at the top. M: DNA molecular maker.
Allelic and genotypic frequencies of PRLR and VRTN
The allelic and genotypic frequencies of PRLR gene according to breeds are given in Table 1. Two alleles (A and B) and three genotypes (AA, AB and BB) were presented in all studies pig groups. The most abundant genotype was the heterozygous one. The frequency of expected genotype AA was 0.35 (Landrace), 0.40 (Yorkshire), and 0.17 (Meishan).
Table 1: Allelic and genotypic frequencies of PRLR gene.
| Breeds | Number of pigs | Genotypic frequencies | Allelic frequencies |
χ² |
P-value | ||||
| AA | AB | BB | A | B | |||||
| Landrace | 37 | 0.35 (13) | 0.51 (19) | 0.14 (5) | 0.61 | 0.39 | 0.18 | 0.92 | |
| Yorkshire | 35 | 0.40 (14) | 0.57 (20) | 0.03 (1) | 0.69 | 0.31 | 3.78 | 0.15 | |
| Meishan | 30 | 0.17 (5) | 0.76 (23) | 0.07 (2) | 0.55 | 0.45 | 8.71 | 0.01 | |
Figures in brackets are the number of samples. χ² = chi-square test.
Table 2: Alleles and genotypic frequencies of VRTN gene.
| Breeds | Number of pigs | Genotypic frequencies | Allelic frequencies |
χ² |
P-value | ||||
| QW | WW | Q | W | ||||||
| Landrace | 37 | 0.30 (11) | 0.27 (10) | 0.43 (16) | 0.43 | 0.57 | 7.34 | 0.03 | |
| Yorkshire | 35 | 0.37 (13) | 0.40 (14) | 0.23 (8) | 0.57 | 0.43 | 15.27 | <0.01 | |
| Meishan | 30 | 0.10 (3) | 0.23 (7) | 0.67 (20) | 0.22 | 0.78 | 2.66 | 0.26 | |
Figures in brackets are the number of samples. χ² = chi-square test.
The frequency of allele A was ranged from 0.55 (Meishan) to 0.69 (Yorkshire). The loci were in HWE for Landrace and Yorkshire herds but not for Meishan.
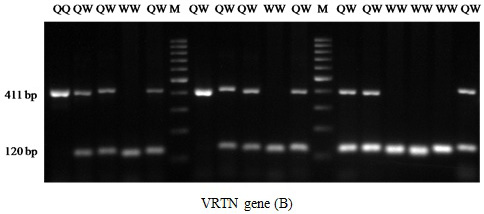
Figure 2: PCR fragments of VRTN separated on 2% agarose gel. The genotypes are shown at the top. M: DNA molecular maker.
For VRTN gene, all pig group are segregating for this mutation. The frequency of QQ was 0.3, 0.37 and 0.1 for Landrace, Yorkshire and Meishan, respectively. The mutant (insert) allele (Q) which associated with more teat and vertebral number (Yang et al., 2016) was ranged from 0.22 (Meishan) to 0.57 (Yorkshire). The genotype frequency distribution of VRTN was in HWE (P>0.05) for Meishan but not for Landrace and Yorkshire (P<0.05).
DISCUSSION
Number of teats is an important trait for breeding programs because the number of piglets in a litter is often larger than the number of functional teats of the sow due to the remarkable improvement in sow prolificacy over the last decades (Rodriguez et al., 2005). A lower number of teats than the number of piglets induces suckling competition, which can lower pre-weaning growth and survival. The total number of thoracic, lumbar vertebrae and ribs are economically important traits that affects production in pigs. A higher of vertebral and ribs number could enhance litter size of sows.
The AluI polymorphism in the porcine PRLR gene and its effects on litter size were studied first by Vincent et al. (1997) and Rothschild et al. (1997). A new primers were designed from the 3′ region of the porcine PRLR cDNA sequence (GenBank accession no. U96306) by Drogemuller et al. (2001). This study established a PCR-RFLP genetic polymorphism at the PRLR locus followed the previous report of Drogemuller et al. (2001). Previous studies refered to the effects of PRLR gene on litter size traits of sows, in our study, the polymorphism of PRLR gene is considered as candidate gene for teat and vertebral number.
Two alleles A and B and three genotypes (АА, АВ and ВВ) were found in all studies population (Landrace, Yorkshire and Meishan). The expected frequency of genotype AA, which is supposed to be positively associated with litter size (Vincent et al., 1998; Drogemuller et al., 2001; Terman, 2005; Omelka et al., 2008) and teat and vertebrate number (Wang et al., 2011; Yang et al., 2016) in pigs was 0.35 (Landrace), 0.40 (Yorkshire) and 0.17 (Meishan). The most abundant genotype was the heterozygous one. The expected frequency of expected allele A was quite high in Landrace, Yorkshire and Meishan herds. Previous studies demonstrated the presence of two alleles A and B and three genotypes (АА, АВ and ВВ) with varying frequencies in the different pig populations. Mihailov et al. (2014) found PRLR allele frequencies of allele A was 0.33, 0.58 and 0.52 and allele B was 0.66, 0.41 and 0.47 in Large White, Landrace and crossbred sows, respectively. In the study of Tempfli et al. (2015) in crossbred mangalica pigs, the similar frequencies of alleles A and B were found (0.52 and 0.48), with relatively lower frequencies of homozygous AA and BB genotypes (0.18; 0.15) and comparatively higher frequency of the heterozygous AB genotype (0.67). Study of Balogh et al., 2016 showed higher frequency for allele A in Hungarian Large White and Pietrain breeds (0.63 and 0.59, respectively) than for allele B frequency (0.37 and 0.41) but in Duroc sows, allele A frequency was lower than that of allele B (0.17 vs 0.83). In our study, a lower frequencies of homozygous AA and BB genotypes as compare with heterozygous AB genotype in all populations; a higher frequencies of A allele in Landrace, Yorkshire and Meishan population than B allele were found.
The VRTN gene appeared as the most promising candidate in the region of SSC7 which has been identified as a QTL for number of teat in pig populations (Ding et al., 2009; Guo et al., 2008; Sato et al., 2006; Wada et al., 2000; Zhang et al., 2007) as well as a QTL for number of vertebrae and ribs (Edwards et al., 2008; Choi et al., 2011; Mikawa et al., 2005; Ren et al., 2012; Sato et al., 2003). VRTN encodes a potential DNA binding factor and has been described as an essential factor for development of the embryo in different species (Mikawa et al., 2011). Due to its biological function, this gene has been indicated as a candidate gene for number of vertebrae (Fan et al., 2013; Mikawa et al., 2011; Ren et al., 2012).
Under this study, all breeds including Landrace, Yorkshire, and Meishan are segregating for the mutation of VRTN gene. The frequency of mutant (insert) allele Q which associated with more vertebral number was similar to allele W in Landrace, Yorkshire pig populations while it was lower than allele B in Meishan. The frequency of expected genotype QQ was low in Meishan herd as compare to other population. This result is consitent with previous study of Yang et al. (2016) who indicated that the mutant (ins) allele predominantly exist in European commercial breeds (Large White: 0.65; Landrace: 0.82). In our study, the frequencies of allele Q (0.43; 0.22) were lower than allele W (0.57; 0.78) in Landrace and Meishan populations.
CONCLUSION
The polymorphic sites of PRLR and VRTN genes were detected in all study populations. For PRLR-AluI, expected frequency of genotype AA was 0.35 (Landrace), 0.40 (Yorkshire) and 0.17 (Meishan), while the frequencies of allele A were 0.61 (Landrace), 0.69 (Yorkshire) and 0.59 (Meishan). In the case of VRTN those values for QQ genotype were 0.30 (Landrace), 0.37 (Yorkshire), and 0.10 (Meishan), while Q allele frequencies were 0.43 (Landrace), 0.57 (Yorkshire) and 0.22 (Meishan).
Authors Contribution
The authors would like to express their deep gratitude to the manager of Thuy Phuong Pig research and Development Center, Vietnam National Institute of Animal Science for providing the pig breeds.
Conflict of interest
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES






