Advances in Animal and Veterinary Sciences
Research Article
First Isolation and Molecular Identification of Alternaria alternata from Camels’ Lung lesion in Egypt
Ashraf A. Abd El Tawab1, Fatma I. El-Hofy1, Amira Mohamed Rizk1*, Azza Said Goda2, Amani Ahmed Hafez2, Shimaa Mohammed Saeed2
1Bacteriology, Immunology and Mycology Department, Faculty of Veterinary Medicine, Benha University, Egypt; 2Animal Health Department, Desert Research Center, Egypt.
Abstract | Purpose: Identification of the causative agent of granulomatous lesion incamel’s lung in Matrouh,Egypt.
Methods: Fifty samples of male camels’ lung from Matrouh abattoir, Egypt were Macroscopically examined for granulomatous lesion. Followed by routine mycological culturing and microscopicl examination of the affected camel’s lung. DNA extraction of the fungal colonies and PCR were done using the Internal Transcribed Spacer (ITS) primers then electrophoresis of the amplified DNA. The 28S ribosomal RNA gene were sequenced followed by alignment on the gene bank data in comparison with previously deposited data of the related fungal sequences. Results: Macroscopic examination of all camels’ lung revealed eight camels (16%) having granulomatous lesions in the lung. Routine mycological examination of these camel’s lung revealed Aspergillus and Alternaria spp. to be the causative agent in 4 of the isolates with a prevalence of (8%). Presence of fungi being the causative agent of the lesion was confirmed by PCR amplification of the ITS region. The species specific analysis of the 28S gene revealed Aspergillus flavus and Alternaria alternata to be the causative agents of the lung lesions. Conclusion: The granulomatous lesion in the camels’ lung were caused by mixed infection by Aspergillus flavus and Alternaria alternata. For the best of the authors’ knowledge, this is the first isolation of Alternaria alternata from camel lung lesion. Thus Alternaria alternata should be considered a potential pathogens not only for animal external lesions but also for internal organs infections
Keywords | Alternaria alternata, Lung, Camel, Molecular, Identification
Received | June 25, 2019; Accepted | August 19, 2019; Published | January 03, 2020
*Correspondence | Amira Mohamed Rizk, Bacteriology, Immunology and Mycology Department, Faculty of Veterinary Medicine, Benha University, Egypt Email: [email protected]
Citation | Abd El-Tawab AA, El-Hofy FI, Rizk AM, Goda AS, Hafez AA, Saeed SM (2020). First isolation and molecular identification of alternaria alternata from camels’ lung lesion in egypt. Adv. Anim. Vet. Sci. 8(1): 89-95.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.1.89.95
ISSN (Online) |2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Rizk et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
For pastoral community, Camel is considered a key player of the subsistence economy in varying zones in the world (Abbas and Omer, 2005). Camel exports have a significant contribution of the national income of the camel-producing countries as Sudan and Somalia (Clarke 1985).They are considered as an important sport animals in the Arabian Gulf countries (Snow et al., 1992). Camels are also used for meat, milk, wool, as well as for transportation purposes (Badawy et al., 2013) in the recent years, being the cause of morbidity and mortality, fungal infection have received a significant concern especially in immunocompromised patient (Low and Rotstein, 2011). Previously, camels were considered to be resistant to most of livestock disease but recently they are considered to be as susceptible as other animals (Abbas and Omer, 2005). Fungal infection occurs in favorable conditions like immunodeficiency, antibacterial use for long time, organ transplant and chemotherapy (Pfaller et al., 2006). Aspergillosis is a mycotic affection caused by Aspergillus species (Chabi et al., 2015). Macroscopically Camel internal Aspergillosis was reported to be caused by Aspergillus fumigatus that has been isolated from internal organs including lungs of racing camels due to fatal respiratory and enteric clinical signs and symptoms. (El-Khouly et al.,1992). Growth and multiplication of this filamentous fungus is stimulated by heat, moisture and organic matters (Chabi et al., 2015). In the last decade, many fungi including Aspergillus flavus have been emerged as the causative agent of morbidity andmortality in humans and animals (Nucci and Marr, 2005, Pal et al., 2014). Previously, Aspergillus flavus was considered to be the second leading cause, after Aspergillus fumigatus, of invasive aspergillosis (Denning, 1998). In Africa, pulmonary aspergillosis was reported to be caused mainly by Aspergillus flavus (Mahgoub and El-Hassan, 1972).
Alternaria is a hyphomycete that is often encountered in human fungal infection (Pastor and Guarro, 2008). This fungus can be encountered on normal animal and human skin (Avsever et al., 2017). Alternaria has been frequently linked with ocular infections, paranasal sinusitis, allergic rhinitis and bronchial asthma (Lyke et al., 2001, Shugar et al., 1981, Stark et al., 2005; Bush and Prochnau 2004). Recently, Alternaria alternate has been isolated and identified from severe fatal lung lesions occurring in the wild Egyptian mole rat (Tamam and Refai, 2013). The aim of this study was to screen for the fungal causes of some granulomatous lesion usually encountered in one humped camels at the slaughter house in Marsa Matrouh Province, North of Egypt.
Materials and Methods
Collection of Samples
The whole lungs were taken out from 50 slaughtered one humped camels (4 to 9 years old) in the period just after slaughtering during early morning. The sampling period extended from December 2015 to June 2016 at main abattoirs in Matrouh province 500 km west of Cairo where camel rearing is common. Of these post slaughtered examined 50 camel, Eight Lungs with macroscopic lesions were obtained. Then, these Lungs were aseptically sampled for mycological, and molecular identification. Sampling was carried out after lungs external surfaces were prewashed using PBS solution and sterilized with 95% ethanol. Using a heat-sterilized scalpel and forceps, all specimens were cut and divided into the required sample for each investigation protocol and transferred to Mycology Research Lab. Desert research center, Egypt (Lemos et al., 2007)
Isolation Strategy
The fungi were isolated by the direct plating method according to (Dhingra and Sinclair, 1995) on Sabaroud dextrose agar medium plates (Oxoid, thermo scientific, UK) with Tetracyclin 0.5 gm/L. The plates were then incubated at 28°C±2 for 5-7 days and every single fungal colony was purified, identified and preserved by transferring to a pure slant containing malt extract agar medium and hence stored at 4°C (Ayman et al., 2017).
Morphological Identification
Isolates were identified on the basis of routine cultural characteristics and morphological characteristics according to (McClenny, 2005).
Electron Microscope
Specimens were fixed in 2.5% (v/v) Glutaraldehyde in phosphate buffer solution for 20 min. The fixed specimens were dehydrated through a series of increasing concentrations of ethanol, 10, 20, 30, 50, 70, 90, 95, 100% at 10 minutes for each, with 3 changes at 100% with the use of Acetone as the intermediate fluid. Critical point drying (CPD) was used to prevent collapse in Scanning Electron Microscope (SEM). The samples were sputter-coated with gold using an Emitech K550X coating unit. Then the specimens were examined usingScanning Electron Microscope (SEM). (Alexander et al., 2017).
Molecular Identification
Fungal DNA Extraction and PCR: DNA extraction was carried out by Quick-DNA™ Fungal/Bacterial Microprep Kit (Zymo research #D6007). Then the ITS region was amplified by PCR using ITS1 and ITS4 in the fungal isolates. PCR was done by using Maxima Hot Start PCR Master Mix (Thermo K1051). Briefly 1 μl of forward and reverse ITS primer (20 μM), 5μl of fungal DNA, 25μl of PCR Master Mix (2X) were added and completed to 50μl of nuclease free double distilled water. The mixtures were vortexed and spin down and put in the thermal cycler for initial denaturation at 95°C for 10 min. this was followed by 35 cycles each consisted of denaturation at 95°C for 30 sec., annealing at 57°C for 1 min, extension at 72°C for 1.5 min. and final extension at 72°C for 10 min.
Horizontal Gel Electrophoresis
PCR-amplified products were separated by agarose gel electrophoresis using a horizontal submarine gel system (AgagelMedi, Biometra) as well as agarose (Gibco BRL Life Technologies) at a concentration of 2% (w/v). Electrophoresis was conducted in 0.5× TBE( Tris borate EDETA) buffer at 10 v/cm for various times, depending on the size of the gel unit, DNA size marker was used as standard (Gene Ruler 100 bp plus DNA ladder; Fermentas Biotech. Inc.).The Run was continued for 1 h. and then agarose gel with DNA bands were stained with ethidium bromide (10 mg/ml) then visualized and photographed under UV light using a camera.
Sequencing of ITS gene:The PCR product was undergo cleaning up using Gene JET™ PCR Purification Kit (ThermoK0701) according to the manufacture instructions. The sequencing of the PCR product was made on GATC German Company using ABI 3730xl DNA sequencer by ITS1 and ITS4 primers in addition to 28S ribosomal RNA gene partial sequences. The DNA sequence was determined by automated DNA sequencing method. The automated DNA sequencing reactions were performed using Big Dye terminator ready sequencing kit. The reaction was conducted in a total volume of 20 µl, containing 8 µl of terminator ready reaction mix, 1 µg of DNA, and 3.2 pmole of forward primer. The cycle sequencing program was 96 °C for 10 seconds, 50°C for 5 seconds, and 60 °C for 4 minutes, repeated for 25 cycles with rapid thermal ramping. The nucleotide sequence was determined automatically by the electrophoresis of the cycle sequencing reaction product on ABI 3730xl DNA sequencer. The data were provided as fluorimetric scans from which the sequence was assembled using the sequence analysis software.
Alignment Analysis
The obtained nucleotide sequences of ITS genes of 4 fungal isolates in addition to 28S ribosomal RNA gene partial sequences were analyzed using GenBank database25 by using BLAST program available on the National Center of Biotechnology Information website (www.ncbi.nlm.nih.gov), where they were compared to the sequences on library in order to assess the gene similarities.
Data Analysis
The sequence data were assembled and analyzed with the help of Sigma package of sequence analysis software (DNAStar Inc., Madison, WI, USA). Sequence similarity check (Blast) were taken out by comparing the sequence to related microbial sequencing in the database (http://www.ncbi.nlm.nih.gov/BLAST/). Related microbial sequence alignments were performed by ClustalX and phylogeny was determined by neighbor-joining method. Prevalence of fungal infections were presented as mean ± SD.
Results
Macroscopic Appearance of the Suspected Fungal-Induced Lung Lesion
Naked eye examination of abnormal lungs of slaughtered camels revealed lungs with white to yellowish granulomas of varying size and multiple abscesses on its caudal lobe containing large amount of fetid grey purulent material (Figure 1).
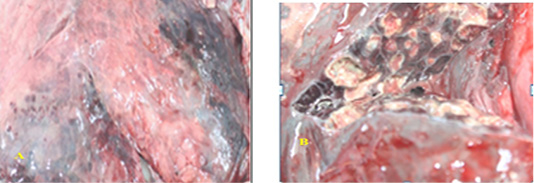
Figure 1: SGranulomatous lesions on the lateral (A) and medial (B) surfaces of camel lung which showing fetid grey purulent material.
Culture Characteristics
Culture characteristics examination by naked eye (Figure 2) revealed velvety surface, olive green color which turns yellowish in old culture with plenty spore formation and slightly pale brown on the reverse side of the colonies, indicating Aspergillus while other culture has cottony colonies, covered by grayish, short, aerial hyphae. The surface is greyish white at the beginning which later darkens and becomes black or olive brown with a light border indicating the possibility of being Alternaria.
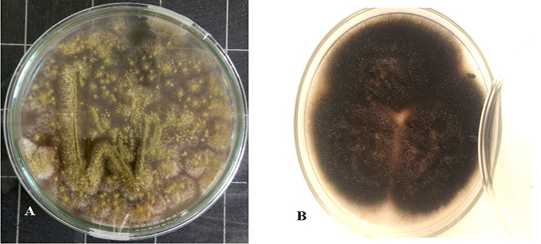
Figure 2: Aspergillus Flavus culture with velvety surface, olive green color(A) and Alternaria alternata culture cottony grey to olive brown colonies on the surface with short aerial hyphae (B).
Microscopic Appearance
Light microscopic examination: revealed some heavy walled, coarsely roughened conidiophores. Vesicles are elongate when young, later becoming subglobose or globose. Phialides are uniseriate. Conidia are typically globose to subglobose indicating to Aspergillus flavus (Figure 3 A). The other conidia are brown in color, ovoid or obclavate, with an elongated apical celltypically in chains. Conidiophores are septate (transverse & longitudinal septations), simple or branched indicating to Alternaria species (Figure 3 B).
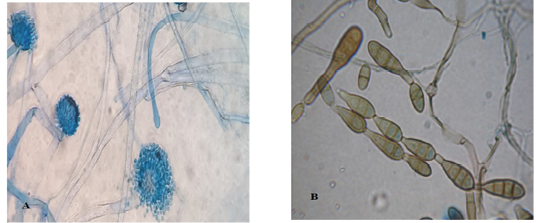
Figure 3: Light microscope examination shows heavy walled, coarsely roughened conidiophores and globose to subgloboseconidiaindicating to A. flavus (A). Other conidia are brown in color with an elongated apical cell typically in chains and Conidiophores are septate indicating to Alternaria species (B).
Electron Microscope Examination
A highly magnified photo by SEM of A. flavus shows conidiophores ends with small head bearing uniseriate phialides terminates with a tight chains of conidia which are globose (Figure 4 A & B). While A highly magnified photo by SEM of Alternaria alternata shows simple unbranched and branched conidial chains of moderate length as well as solitary conidia are interspersed across the colony especially areas that receive little light conidia of A. alternata which tends to grow in longer chains and have a median and subconstricting transverse septum dark brown conidia with short beaks Figure 4 (C & D).

Figure 4: SEM of A.flavus A and B showing 10 µm,1000x and 2000x a highly magnified photo for the shape of conidiophores which ends with small head bearing uniseriatephialides terminates with a tight chains of conidia which are globose. C and D show SEM of 20 µm 850x magnified and 10um 2.300 x of Alternaria alternata with simple unbranched and branched conidial chains of moderate length
Molecular Identification
The ITS is considered the barcode for identification of fungal DNA. The DNA complete sequences of Aspergillus flavus using ITS primer set 1 and 4 produced the gene amplified size of 1141 (bp).
The ribosomal RNA gene is encoded by DNA coding sequences. In fungus this (rDNA) consists of genes encoding the 18S, 5.8S and 28S rRNA subunits which are encompassed by ITS1; ITS2. The 18S-ITS1-5.8S-ITS2-28S DNA region that is ideal as a target for molecular characterization. The alignment of Aspergillus flavus ITS sequences revealed 99% identity with the published Aspergillus flavus ITS in the gene bank library
ITS gene sequences was used to confirm the phylogenic relationship of the obtained sequences with Aspergillus species resulted in the appearance of identity of the sequence to a degree of 99% with published Aspergillus Flavus sequences (Figure 5).The alignment clarified that the specific gene is resemble toA. flavus by 99% which support the morphological identification

Figure 5: Dendrogram of A. flavus isolate showing its identity with public library. The amplified gene 1141 (bp) was used to insert the organism in its position in the phylogenetic tree.
The ITS is considered the barcode for identification of fungal DNA. The DNA complete sequences of Alternaria alternata using ITS primer set 1 and 4 produced the gene amplified size of 551 (bp).
The ribosomal RNA gene is encoded by DNA coding sequences. In fungus (rDNA) consists of genes encoding the 18S, 5.8S and 28S rRNA subunits which are encompassed by ITS1; ITS2. The 18S-ITS1-5.8S-ITS2-28S DNA region that is ideal as a target for molecular characterization fig. The alignment of Alternaria alternata ITS sequences revealed 99% identity with the published Alternaria alternata ITS in the gene bank liberary
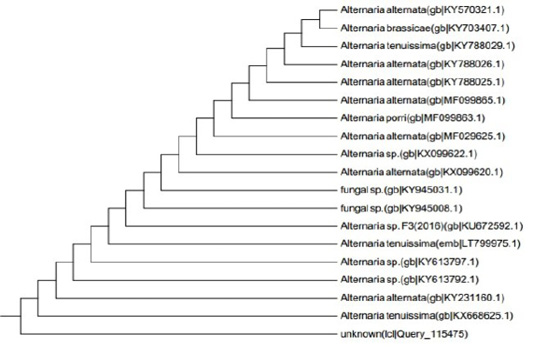
Figure 6: Dendrogram of Alternaria alternata isolate showing its identity with public library. The amplified gene 551 (bp) was used to insert the organism in its position in the phylogenetic tree.
ITS gene sequences was used to confirm the phylogenic relationship of the obtained sequences with Aspergillus species resulted in the appearance of identity of the sequence to a degree of 99% with published Alternaria Alternata sequences (Figure 6) the alignment clarified that the specific gene is resemble toAlternaria alternata by 99% which support the morphological identification
Discussion
Fungal infections is considered to be an important cause of morbidity and mortality especially in the immune compromised animals (Chabi et al., 2015). Exposure to antibacterial, chemotherapy or even immunosuppression for extended period are known to predispose for high fungal infection (Pfaller et al., 2006). Although Alternaria was reported to be encountered on normal human and animal skin (Yu, 1965), but it was also the causative agent of allergic sinusitis (Shugar et al., 1981), rhinitis (Pant et al., 2005), bronchial asthma (Bush and Prochnau 2004) and hypersensitivity pneumonitis (Ogawa et al., 1997).
In the current study, mycotic examination of granulomatous lesion in camel-lung revealed Alternaria alternata that could be isolated and identified in association with Aspergillus flavus. Although pulmonary Alternaria-induced granulomatous lesion is rare, granulomatous lung lesion were reported to be caused by Alternaria alternata in human (Lobritz et al., 1979).
The light and electron microscopic investigation of the causative agent of the encountered granulomatous lesion in the camel lung in this study revealed the presence of fungal characteristics indicating to Alternaria species and Aspergillus spp. The isolation and identification of Aspergillus flavus from granulomatous lesion of camel lung may be unusual. The molecular identification by fungal sequencing and analysis beside phylogenic analysis of the fungal isolates in the current work revealed Aspergillus flavus and Alternata alternaria in the same lung lesion in camel which led us to believe that the lesion is caused by a mixed infection of both microorganism.
At risk for fungal infection include also normal individual as cases of invasive aspergillosis have been reported in normal hosts after extensive environmental exposure to Aspergillus spores in the form of tree-bark chippings (Butler et al., 2013), or after inhalation of a great deal of dust in a mushroom factory (Shimaoka et al., 2006). Considering the desert nature of the sampling area and the continuous windy environment in the current study may explain the chance of getting pulmonary fungal infection in some weak camel that are not sheltered all the time.
Internal transcribed spacer (ITS) region sequences analysis has been proved to be validated for fungal spp identification (deHoog and Horré 2002). It also was chosen as the official barcode for fungi identification by a consortium of mycologists (Schoch et al., 2012). Aspergillus flavus has been reported to cause allergic rhinosinusitis and fungal keratitis (Kosmidis and Denning 2015).Worldwide, Aspergillus flavus has been reported to be the second after Aspergillus fumigatus, most frequent causative agent of invasive Aspergillosis (Foshee et al., 2016). However, invasive aspergillosis in some tropical and subtropical countries was reported to be predominantly caused by Aspergillus flavus (Zarrinfar et al., 2012).This is consistent with the encountered Aspergillus flavus being the causative agent pulmonary lesion in this study.
Mycotic pneumonia has been also reported to be caused by Aspergillus species (Carlos et al., 2012). Aspergillus fumigatus was considered to be the most common cause associated with Aspergillus pulmonary infection meanwhile Aspergillus flavus was considered the common cause of allergic rhinosinusitis, postoperative aspergillosis and fungal keratitis (Kosmidis Denning, 2015).
The air borne disease Alternariosis is caused by Alternaria alternata can occurs in animals and humans (Andersen et al., 2001). Besides being the causative agents of, ulcerated cutaneous infections, and keratitis, Alternaria alternata has been recently reported to be the causative agent of Allergic fungal sinusitis (Abdolrasoul et al., 2017). Alternaria alternata has been also reported to be the causative agent of visceral infections and osteomyelitis (Anaissie et al., 1989, Manning et al., 1991). Additionally Alternaria alternata has been reported to be among the causative agents of otitis media in agricultural field workers (Wadhwani, and Srivastava, 1984).Therefore the granulomatous lesion found in the camel lung in our study is most probably caused by mixed infection by Aspergillus flavus and Alternaria alternata.
Conclusion
The causative agents encountered in granulomatous lung lesion in one humped camel in the desert area in the north of Egypt is most probably caused by mixed infection by Aspergillus flavus and Alternaria alternata. Moreover, Alternaria alternata should be considered a potential pathogens not only for animal external infection but also for internal organs in camel and other animals especially those exposed to harsh environmental conditions.
acknowledgements
We thank professor Dr. Waleed Bakry, professor of mycology, faculty of science, Al-Azhar University, Egypt for support in processing of samples
Conflict of Interest
The authors report no conflicts of interest. The authors alone were responsible for the present paper.
authors contribution
Professor Ashraf A. Abd El TAwab: shared in the study design, development of methodology and manuscript revision. Dr.Fatma I. El Hofy: shared in the study design, development of methodology, manuscript revision. Dr. Amira Mohamed Rizk: Shared in the study design, development of methodology, collection of data, data analysis and interpretation in addition to writing and revision of the manuscript. Professor Azza said Goda:Shared in the study design, development of methodology, collection of data. Dr. Amani Ahmed Hafez: shared in the study design, development of methodology and collection of data. Mrs Shimaa Mohammed saeed: sample collection, methodology and writing of the manuscript.
References






