Advances in Animal and Veterinary Sciences
Research Article
Enzyme Activity Assay of Lactic Acid Bacteria from Civet (Paradoxurus hermaphroditus) Digestive Tract
Fitri Fitri1, Abu Bakar Tawali2*, Amran Laga2, Zaraswati Dwyana3
1Agricultural Science Study Program, Postgraduate School of Hasanuddin University, Perintis Kemerdekaan Street Km. 10 Tamalanrea, Makassar, 90245, Indonesia; 2Food Science and Technology Study Program, Department of Agricultural Technology, Faculty of Agriculture, Hasanuddin University, Perintis Kemerdekaan Street Km. 10 Tamalanrea, Makassar, 90245, Indonesia; 3Department of Biology, Faculty of Math and Science, Hasanuddin University, Perintis Kemerdekaan Street Km. 10 Tamalanrea, Makassar, 90245, Indonesia.
Abstract | Food and non-food fermentation actually use lactic acid bacteria alteration as agent in the production process for enhance the aroma and taste of the product, in the case of civet coffee. This study aimed to determine and identified the ability of enzymes produced by lactic acid bacteria from civet digestion tract. The enzymes analyzed in this study were protease, lipase, and cellulase enzymes. The activity of protease carried out by the Lowry method, the lipase activity used the titrimetric method, while the cellulase activity used dinitrosalicylic (DNS) colorimetric method. The result showed that lactic acid bacteria from civet can produced protease, lipase, and cellulase on different capacity. For the protease enzyme, the highest activity enzyme was produced by Leuconostoc pseudomesenteroides Ni1324 (0.051 U/mL), while the lowest was produced by Weissella cibaria MG5327 (0.041 U/mL). For the lipase enzyme, the highest activity enzyme was produced by Leuconostoc pseudomesenteroides Ni1324 (0.639 U/mL), while the lowest was produced by Weissella cibaria MG5327 (0.306 U/mL). And for the cellulase, the highest activity enzyme was produced by Weissella cibaria MG5327 (0.039 U/mL), while the lowest was produced by Leuconostoc pseudomesenteroides CF102 (0.029 U/mL)
Keywords | Lactic acid bacteria, Enzyme activity, Protease, Lipase, Cellulase
Received | June 10, 2021; Accepted | June 16, 2021; Published | August 25, 2021
*Correspondence | Abu Bakar Tawali, Food Science and Technology Study Program, Department of Agricultural Technology, Faculty of Agriculture, Hasanuddin University, Perintis Kemerdekaan Street Km. 10 Tamalanrea, Makassar, 90245, Indonesia; Email: [email protected]
Citation | Fitri F, Tawali AB, Laga A, Dwyana Z (2021). Enzyme activity assay of lactic acid bacteria from civet (paradoxurus hermaphroditus) digestive tract Adv. Anim. Vet. Sci. 9(10): 1649-1654.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.10.1649.1654
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Tawali et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Lactic acid bacteria (LAB) are Gram-positive bacteria without cytochromes that have a rod or round shape, non-sporing, facultatively anaerobic, cannot reduce nitrates, negative-oxidation, negative-catalase, where lactic acid fermentation is the primary metabolism (Temmerman et al., 2004; Bintsis, 2018). Lactic acid bacteria are commonly used in the food and non-food industries due to their ability to generate a variety of enzymes. In the food industry, lactic acid bacteria are widely used to produce various fermentation products (Rhee et al., 2011). In addition, lactic acid bacteria also play a role in increasing the aroma and taste of food products, for example in coffee (Pereira et al., 2020) and yogurt making (Chen et al., 2017).
Lactic acid bacteria groups are found in many fermented products (Vasiljevic and Shah, 2008). In addition, the lactic acid bacteria group also lives in the gastrointestinal and urogenital tracts of humans and animals (Vasiljevic and Shah, 2008; Aureli et al., 2011), including in the digestive tract of civet (Suhandono et al., 2016). Civet (Paradoxurus hermaphroditus) is a mammal widely cultivated in Indonesia to produce luwak coffee, one of the world’s most expensive coffees originating from Indonesia (Winaya et al., 2020; Schmidt-Burbach et al., 2014). This coffee has a unique taste caused by the fermentation of bacteria in the digestive tract of civet.
Fermentation is one of the crucial processes in forming the taste and aroma of the coffee beans. The components contained in coffee beans are degraded by enzymes produced by bacteria during fermentation. Elements such amino acids, peptides, sugars, fatty acids, and organic acids can affect the quality of a coffee bean (Lee et al., 2015).
The special of the aroma and taste of luwak coffee has led to many studies developing lactic acid bacteria from civet digestive for coffee fermentation (Rubiyo and Towaha, 2013; Hadipernata, 2018). Although the exploration of lactic acid bacteria from civet digestive has begun to be developed, information regarding these bacteria’s enzyme activity is still not widely found. Therefore, this study was conducted to determine the enzyme production of lactic acid bacteria isolated from civet digestion. This is expected to be additional information in developing the use of lactic acid bacteria from civet in enhancing the aroma and taste of coffee.
Material and Methods
Material
Five species of lactic acid bacteria were used in this study, namely Lactobacillus plantarum CAU:227, Lactobacillus plantarum IMAU20905, Weissella cibaria MG5327, Leuconostoc pseudomesenteroides CF102, and Leuconostoc pseudomesenteroides Ni1324. These isolates were obtained from luwak digestion from previous studies (Tawali et al., 2019).
Methods
Production of Crude Protease from LAB
Crude protease extract from lactic acid bacteria was carried out in 250 ml of media with the following composition: 1.88 gr peptone; glucose 1.25 g; 125 ml mineral salt (MgSO4 0.1 g; KH2PO4 0.1 g; FeSO4 0.02 g). As much as 2% of each isolate that had been rejuvenated on MRS Broth media piped into the production medium. These isolates incubated using a shaker at 120 rpm for 18 hours. Then, centrifuge the sample at 12000 rpm at 4oC for 15 minutes. The supernatant, which is the crude extract of the enzyme, has been taken and used for the enzyme assay.
Production of Crude Lipase from LAB
The crude lipase extract was produced in 250 ml of MRS Broth media enriched with 1% olive oil. As much as 2% of each rejuvenated isolate was pipetted into the production medium. Incubation was carried out using a shaker at a speed of 120 rpm for 18 hours. Crude extract of the enzyme was obtained by doing centrifugation at 12000 rpm at 4oC for 15 minutes. Then, the filtrate was separated from the supernatant, and the supernatant was taken as crude extract of the lipase enzyme.
Production of Crude Cellulase from LAB
Production of bacterial cellulase crude extract was carried out on 250 ml of media with the following composition: CMC 2.5 gr; yeast extract 0.5 gr; glucose 0.25 g; MgSO4 0.05 gr; KNO3 0.187 gr; K2HPO4 0.125 gr; FeSO4 0.005 gr; CaCl2 0.01 gr. As much as 2% of the rejuvenated isolates on MRS Broth media were pipetted into the production medium. Then, incubated using a shaker at a speed of 120 rpm for 18 hours. After the incubation period is complete, centrifugation is carried out at a speed of 12000 rpm at 4oC for 15 minutes. The supernatant, which is the crude extract of the enzyme, is then taken and used for the enzyme assay step.
Protease Activity Assay of LAB
Protease enzyme activity assay used in this research was a method from Sony and Potty (2016) that has been modified. As much as 0.2 ml of the sample was pipette and added 1 ml of Tris HCl buffer solution pH7. After that, 1 ml of 20 mg/ml casein solution was added and incubated at 37oC for 30 minutes. Then 1 ml of TCA solution was added to stop the enzyme activity. Next, each sample was piped as much as 1.5 ml, and 2.5 ml of Na2CO3 was added. Then added 1 ml of Folin-Ciocalteu 50% reagent. It was allowed to stand for 30 minutes and the absorbance at 670 nm wavelength. For control, the same steps are carried out as in the sample, except that the incubation process is not carried out. The protease enzyme activity value is then calculated using the formula:
Enzyme Activity (U/mL) =
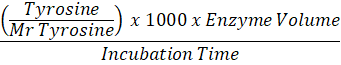
Lipase Activity Assay of LAB
Lipase activity assay used in this research was a method from Indriawan et al. (2018) that has been modified. Weighed 1 gram of palm oil and then added a sample of 0.5 ml. As much as 2 ml of pH 7.5 phosphate buffer was added and then stirred for 1 hour at 37oC. After that, add 5 ml of acetone: alcohol (1: 1). Then three drops of pp indicator were added and titrated using KOH-alcoholic 0.05 N. The same procedure was carried out for control by replacing palm oil with distilled water. The lipase activity is then calculated using the formula:
Enzyme Activity (U/mL) =

Cellulase Activity Assay of LAB
Cellulase activity assay used in this research was a method from Dini and Munifah (2014) that has been modified. As much as 0.5 ml of sample was added with 0.5 ml of phosphate buffer pH 7 and 1% CMC solution, homogenized using a stirrer. After that, 1.5 ml of DNS reagent was added and incubated at 37oC for 30 minutes. It has been heated using a hotplate for 10 minutes and then cooled. Measurements were made using a spectrophotometer at 470 nm wavelength. Cellulase activity is then calculated using the formula:
Enzyme Activity (U/mL) =
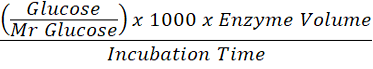
Data Analysis
The research data were analysed using One-Way ANOVA with three replication. Tuckey test at a confidence level of 0.05 (p <0.05) was used to see the difference in each sample.
Result and Discussion
Protease Enzyme Assay of Lactic Acid Bacteria
Protease enzyme activity assay in this study was carried out using the principle of the Lowry method. The substrate used in this study was casein. Casein which is hydrolyzed by protease, will release tyrosine and other amino acids. The released tyrosine can react with the folin reagent (Cupp-Enyard, 2008). This reaction will produce a blue colour whose colour density depends on the tyrosine concentration contained in the sample. The advantages of the Lowry method are sensitivity, specificity, and less affected by sample turbidity (Pratama, 2019).
The tyrosine standard curve was created before the protease activity assay. In making standard curves, the concentration of tyrosine solution was varied to 0.05; 0.1; 0.2; 0.4; 0.8 mg / mL, resulting in a linear equation y = 1.9759x + 0.0351 with a regression coefficient value (R2) of 0.9987 (Figure 1). The standard curve serves to determine the concentration of tyrosine from hydrolysis present in the sample.
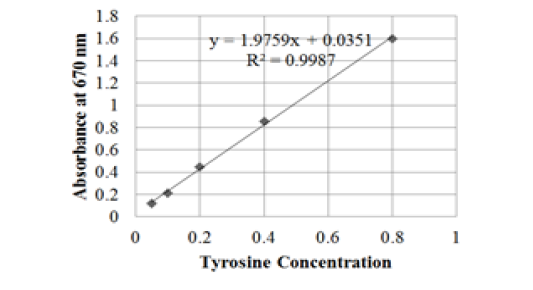
Figure 1: Tyrosine standard curve
The results of lactic acid bacteria’s protease activity assay from civet can be seen in Figure 2. Based on Figure 2 it can be seen that all of the isolates obtained from civet were able to produce the protease enzyme. Leuconostoc pseudomesenteroides Ni1324 (0.051 U / mL) produced the highest protease activity, while the lowest protease activity was produced by Weissella cibaria MG5327 (0.041 U / mL).
Protease is an enzyme that plays a role in protein breakdown reactions. This enzyme can hydrolyze peptide bonds in proteins to produce peptides and amino acids (Naidu, 2011). Peptides and amino acids are essential aroma precursors in the formation of aroma and taste in coffee beans (Poisson et al., 2009).
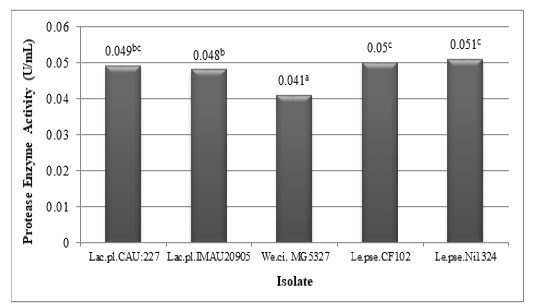
Figure 2: Protease enzyme activity of lactic acid bacteria (Lactobacillus plantarum CAU:227, Lactobacillus plantarum IMAU20905, Weissella cibaria MG5327, Leuconostoc pseudomesenteroides CF102, and Leuconostoc pseudomesenteroides Ni1324) isolated from civet (different superscripts indicate significant differences)
Lipase Enzyme Activity Assay of Lactic Acid Bacteria
The lipase enzyme activity assay was carried out using the titrimetric method. This method is the most common method used in lipase enzyme testing because of its reproducibility, accuracy, and simplicity (Gupta et al., 2003). The principle of testing this method is to measure the free fatty acid content resulting from hydrolysis by lipase (Lopes et al., 2011).
The results of lactic acid bacteria’s lipase enzyme activity assay can be seen in Figure 3. Based on Figure 3 it can be seen that all lactic acid bacteria obtained from civet digestion are able to produce lipase enzymes. The highest lipase activity was produced by Leuconostoc pseudomesenteroides Ni1324 (0.639 U / mL), while the lowest lipase activity was produced by Weissella cibaria MG5327 (0.306 U / mL).
Lipase is a group of enzymes that generally function in the hydrolysis of triacylglycerols (triglycerides) to produce fatty acids and glycerol (Poedjiadi and Supriyanti, 2009). In roast coffee beans, triacylglycerols are important aroma carriers (Petracco, 2005). The fatty acid (FA) fraction of triacylglycerols develops oxidation byproducts as a result of temperature, and primarily consists of aldehydes that react with Maillard reaction intermediates, imparting flavor and aroma to the coffee (Flament, 2001).
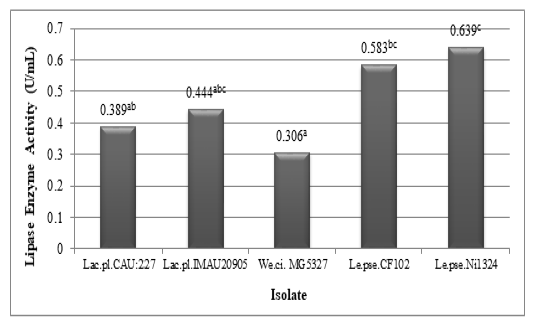
Figure 3: Lipase enzyme activity of lactic acid bacteria (Lactobacillus plantarum CAU:227, Lactobacillus plantarum IMAU20905, Weissella cibaria MG5327, Leuconostoc pseudomesenteroides CF102, and Leuconostoc pseudomesenteroides Ni1324) isolated from luwak (different superscripts indicate significant differences)
Cellulase Enzyme Activity Assay
Cellulase enzyme activity assay in this study was carried out using the DNS method. The principle of quantitative testing for cellulase enzyme activity is to measure the levels of reducing sugars resulting from the enzyme hydrolysis process on the substrate (Lone et al., 2012). Among several cellulase enzyme testing methods, the DNS method is the most widely used because the DNS reagent can measure the resulting reducing sugar levels with a high degree of accuracy, so that it can be used to measure even small groups of sugar concentration (Gusakov et al., 2011).
Reducing sugar levels are determined by preparing a standard glucose curve which functions to determine the concentration of the solution based on its absorbance value. Glucose is used as a solution for making standard curves because glucose is a product resulting from hydrolysis by cellulase enzymes on cellulose substrates (Soeka et al., 2019). Standard curves were prepared using a glucose solution with a concentration of 0.1; 0.2; 0.4; 0.6; 0.8 (mg / mL) to produce a linear equation y = 1.9016x - 0.0011 with a regression coefficient value (R2) of 0.993 (Figure 4). This equation is then used to determine the glucose level from hydrolysis in the sample.
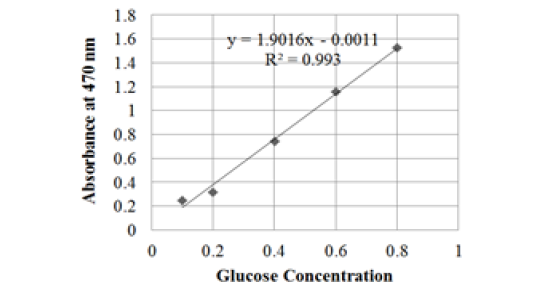
Figure 4: Glucose standard curve
The results of the activity assay for the cellulase enzyme of the lactic acid bacteria isolated from civet can be seen in Figure 5. Based on Figure 5, it can be seen that all isolates obtained from civet are able to produce cellulase enzymes. The highest activity value was produced by Weissella cibaria MG5327 (0.039 U/mL), while the lowest activity value was produced by Leuconostoc pseudomesenteroides CF102 (0.029 U/mL).
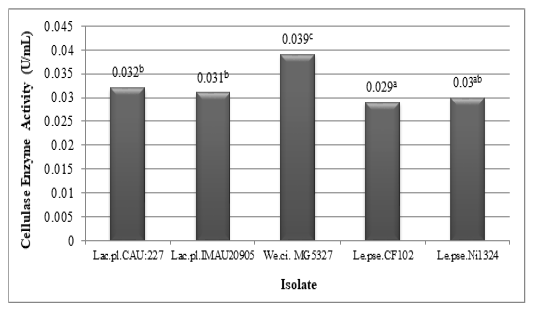
Figure 5: Cellulase enzyme activity of lactic acid bacteria (Lactobacillus plantarum CAU:227, Lactobacillus plantarum IMAU20905, Weissella cibaria MG5327, Leuconostoc pseudomesenteroides CF102, and Leuconostoc pseudomesenteroides Ni1324) isolated from luwak (different superscripts indicate significant differences)
Cellulase are enzymes that can hydrolyze cellulose by breaking the β-1,4 glycosidic bonds in cellulose, cellodectrin, cellobiose, and other cellulose derivatives into simple sugars or glucose (Dini and Munifah, 2014). Reducing sugar is one of the primary precursors in the formation of aroma in coffee. The reaction between amino acids and reducing sugars during the roasting process can produce various volatile compounds in coffee (Lee et al., 2015).
Conclusion
The five isolates of lactic acid bacteria obtained from civet digestion were able to produce protease, lipase, and cellulase enzymes with various enzyme activity values. For the protease enzyme, the highest activity enzyme was produced by Leuconostoc pseudomesenteroides Ni1324 (0.051 U/mL), while the lowest was produced by Weissella cibaria MG5327 (0.041 U/mL). For the lipase enzyme, the highest activity enzyme was produced by Leuconostoc pseudomesenteroides Ni1324 (0.639 U/mL), while the lowest was produced by Weissella cibaria MG5327 (0.306 U/mL). And for the cellulase, the highest activity enzyme was produced by Weissella cibaria MG5327 (0.039 U/mL), while the lowest was produced by Leuconostoc pseudomesenteroides CF102 (0.029 U/mL)
Acknowledgements
This research was funded by Directorate of Research and Community Service, the Ministry of Research and Technology of Indonesia.
conflict of interest
There is no conflict of interest.
authors contribution
All authors contributed equally.
References






