Advances in Animal and Veterinary Sciences
Research Article
Features of Holstein Cattle Bred in Kazakhstan by the Polymorphic Genes of the Somatotropin Cascade
Indira Saltanovna Beishova1*, Vadim Aleksandrovich Ulyanov1, Gulshat Shaikamal1, Natalya Papusha1, Elena Valentinovna Belaya2
1Republican state enterprise on right of economic conducting “Kostanay State University A. Baitursynov” Ministry of Education and Science of the Republic of Kazakhstan, Baytursynov Street, 47, Kostanay, 110000, Kazakhstan; 2Belarusian State Pedagogical University named after Maxim Tank, Sovetskaya st., 18, Minsk, 220050, Belarus.
Abstract | The article presents the results of studying the genetic structure by bGH-AluI and bIGF-1-SnaBI polymorphisms in Holstein cows bred in Kazakhstan. It has been shown that bGH and bIGF-1 genes in the Kazakh population of Holstein cows are polymorphic. By the growth hormone gene, the frequency of bGH-AluIL allele is 0.82; of bGH-AluIV allele – 0.18; by the gene of the insulin-like growth factor-1, the frequency of bIGF-1-SnaBIA allele is 0.58; and of bIGF-1-SnaBIB allele – 0.42. By all the studied polymorphic loci, the Kazakhstan population of Holstein cows is in the state of Hardy-Weinberg genetic equilibrium. The authors believe that research should be continued and the effect of genotypes of bGH-AluI and bIGF-1-SnaBI polymorphisms on milk production should be determined.
Keywords | Holstein breed, Growth hormone gene (bGH), Insulin-like growth factor gene -1 (bIGF-1), Polymorphism, Allele, Genotype
Received | June 12, 2019; Accepted | August 30, 2019; Published | October 15, 2019
*Correspondence | Indira Saltanovna Beishova1, Republican state enterprise on right of economic conducting “Kostanay State University A. Baitursynov” Ministry of Education and Science of the Republic of Kazakhstan, Baytursynov Street, 47, Kostanai, 110000, Kazakhstan; Email: [email protected]
Citation | Beishova IS, Ulyanov VA, Shaikamal G, Papusha N, Belaya EV (2019). Features of holstein cattle bred in kazakhstan by the polymorphic genes of the somatotropin cascade. Adv. Anim. Vet. Sci. 7(s1): 60-65.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.s1.60.65
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Beishova et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Currently, dairy farming is one of the most important branches of agriculture in Kazakhstan. Along with the extensive way of increasing milk production, more and more attention is attached to the intensification of the industry through improving the genetic potential of the bred animals.
Modern achievements in molecular genetics have made it possible to identify the genes associated with qualitative and quantitative traits of cattle. Identification of the preferred allelic variants of such genes will allow breeding with the use of markers at the level of DNA. One of them is the growth hormone gene, as well as the insulin-like growth factor-1.
The growth hormone is involved in regulating growth and development in animals and affects the quality parameters of milk cows. The relationship between polymorphic variants of the growth hormone gene and milk productivity was studied by many foreign scientists. For instance, Grochowska R. and Zwierzchowski L. found a significant correlation between the presence of bGH-AluIL allele and high milk yield of Holstein cows (Grochowska et al., 2001; Zwierzchowski et al., 2002; Chen et al., 2018a, 2018b).
Along with the growth hormone, the insulin-like growth factor-1 regulates growth, development, and lactation. Association of bIGF-1 gene with traits of milk productivity is studied in various countries. For instance, Mehmannavaz et al., 2010 discovered that animals of the Iranian population of Holstein cattle with bIGF-1-SnaBIAB genotype featured high content of fat and protein in milk (Mehmannavaz et al., 2010; Zhang et al., 2018; Beishova et al., 2017).
Siadkowska et al. (2006) also discovered a positive association between bIGF-1-SnaBIAB genotype and a high percentage of milk fat and protein in the Polish population of Holstein cows. Thus, studying the polymorphisms of growth hormone genes and insulin-like growth factor-1 is interesting from the point of determining the genetic potential of cattle by the quantitative productivity traits.
The aim of the study was analyzing the genetic structure by bGH-AluI and bIGF-1-SnaBI polymorphisms in Holstein cows bred in Kazakhstan.
Materials And Methods
The study was performed at the Department of Molecular Genetic Studies of the Research and Innovation Center of the Kostanay State University (KSU) n.a. A. Baitursynov. The object of the study was a group of Holstein cows bred in Kazakhstan (100 animals, LLC Bek+, Kostanay region). The biological material was chosen by the personnel of the farm, and by the personnel of the Department of Molecular-Genetic Studies.
The genotypes of the animal by the polymorphic sections of bGH and bIGF-1 genes were determined using the method of polymerase chain reaction, followed by an analysis of polymorphism of the restriction fragments lengths (PCR-RFLP). DNA was extracted using commercial kit PureLink Genomic DNA Mini Kit. The DNA concentration was measured on spectrophotometer Dynamica Halo DNAmaster. The polymerase chain reaction was performed on the amplifier ProFlex PCR System (Applied Biosystems). The composition of the reaction mixture was the following: water–11.8 µl, 10X buffer–2 µl, dNTP (2.5 mM)–0.4 µl, MgCl2 (25 mM)– 2 µl, primer F (10 pM)–1 µl, primer R (10 pM)–1 µl, and Taq DNA Polymerase (5U/µl)–0.3 µl. The primers sequences and their annealing temperature are shown in Table 1.
The obtained amplificates of bGH and bIGF-1 genes were restricted using the SnaBI and AluI (Thermo Scientific) restriction endonucleases. After incubation, the obtained fragments were separated in 3% agarose gel (Invitrogen). To visualize the results of electrophoresis, the Quantum 1100 gel documentation system (Vilber Lourmat) was used.
bGH-AluI polymorphism is contingent on transition C→G, resulting in amino acid leucine replacement with valine in the protein sequence. The allele recognized by the enzyme contains nucleotide C and is labeled as bGH-AluIL. If the G nucleotide is present, the restriction segment disappears; such an allele is denoted as bGH-AluIV (Figure 1).
Polymorphism of the nucleotide sequence of bIGF-1 gene in area P1 of the promotor region is identified as T→C transversion. Fragment of bIGF-1 gene with the length of 249 b.p. is amplified. The length of fragments after the restriction is 223 and 26 b.p. (Figure 2).
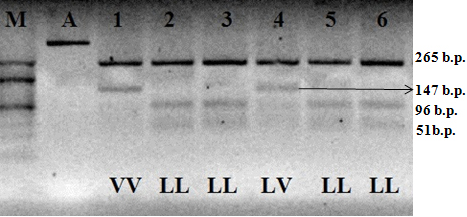
Figure 1: Electrophoretogram of the amplification products and restriction of the fragment of bGH gene; 1–6 hole numbers; A). Amplificate of the polymorphic section of gene bGH; M). The marker of molecular masses O’ RangeRuler 20 bp DNA Ladder (Thermo Scientific); LL, LV and VV–relevant genotypes.
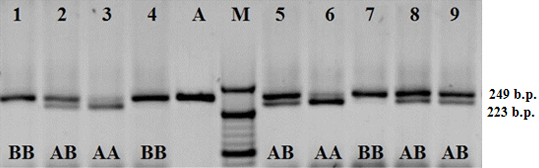
Figure 2: Electrophoretogram of the amplification products and restriction of the fragment of bIGF-1-SnaBI gene; 1–9 hole numbers; A). Amplificate of the polymorphic section of gene bIGF; M) The marker of molecular masses O’ RangeRuler 20 bp DNA Ladder (Thermo Scientific); AA, AB and BB–relevant genotypes.
Based on the obtained data, the expected frequency of genotypes and alleles was calculated according to the genetic equilibrium of the population by Hardy-Weinberg using the χ2 method.
RESULTS AND DISSCUSSION
After the PCR-RFLP analysis, distribution of animals by genotypes of the locus of the growth hormone gene (bGH) was the following: 67 cows had the homozygous bGH-AluILL genotype; 29 cows had the heterozygous bGH-AluILV genotype, and four cows had the homozygous bGH-AluIVV genotype. With that, the frequency of bGH-AluIL allele was 0.82, of bGH-AluIV allele– 0.18 (Table 2).
Many scientists in the near and far abroad countries have analyzed the genetic structure of bGH-AluI polymorphism in various populations of cattle (Table 3). The frequency of occurrence of bGH-AluIV allele in various cattle breeds varies between 0.064 and 0.280 for dairy cattle, and between 0.174 and 0.358 for beef breeds
Table 1: The primers sequence for amplification of polymorphic sections of the growth hormone gene and insulin-like growth factor-1.
| Gene | Primers sequence | Primers annealing temperature | Amplificate size, pairs of nucleotides (b.p.) | Reference |
| bGH |
F: 5′-ccgtgtctatgagaagc-3′; R: 5'-gttcttgagcagcgcgt-3′ |
60° C | 451 b.p. | (Lucy et al., 1993) |
| bIGF-1 |
F:5′-attacaaagctgcctgcccc-3′; R:5′-accttacccgtatgaaaggaatatacgt-3′ |
62° C | 249 b.p. | (Skinkytė et al., 2005). |
Table 2: Frequencies of alleles and genotypes of bGH-AluI polymorphism in the Holstein cattle bred in Kazakhstan.
| Allele | Observed allele frequencies | Relative allele frequencies | Genotype | Number of genotypes | Frequencies of genotypes |
|
bGH-AluIL |
163 | 0.82 ± 0.004 |
bGH-AluILL |
67 | 0.67 |
|
bGH-AluIV |
37 | 0.18 ± 0.004 |
bGH-AluILV |
29 | 0.29 |
|
bGH-AluIVV |
4 | 0.04 | |||
| Total | 100 | 1 |
Table 3: Distribution of the relative frequencies of the alleles of bGH-AluI polymorphism in various populations of Holstein cattle.
| Breed | Relative frequencies of alleles | Frequencies of genotypes | Country | Author, reference | |||
|
bGH-AluIL |
bGH-AluIV |
bGH-AluILL |
bGH-AluILV |
bGH-AluIVV |
|||
| Dairy cattle breeds | |||||||
|
Holstein (n = 134) |
0.936 | 0.064 | 0.87 | 0.13 | 0 | Iran | (Lucy et al., 1993) |
| Holstein | 0.93 | 0.07 | 0.85 | 0.15 | 0 | USA | (Sadeghi, et al., 2008) |
|
Holstein-Friesian (n = 19) |
0.92 | 0.08 | 0.84 | 0.16 | 0 |
New Zealand |
(Hartatik et al., 2016) |
|
Holstein-Friesian (n = 43) |
0.90 | 0.10 | 0.79 | 0.21 | 0 | Australia | (Hartatik et al., 2016) |
|
Black and White (n = 250) |
0.72 | 0.28 | 0.195 | 0.625 | 0.180 | Russia | (Dolmatova and Ilyasov, 2011) |
| Beef cattle breeds | |||||||
|
Kazakh white-headed (n = 296) |
0.826 | 0.174 | 0.67 | 0.31 | 0.02 | Kazakhstan | (Beishova, et al., 2018) |
|
Angus (n = 116) |
0.681 | 0.319 | 0.102 | 0.434 | 0.464 | Ukraine | (Fedota, et al., 2016) |
|
Auliekol (n = 284) |
0.667 | 0.333 | 0.45 | 0.44 | 0.11 | Kazakhstan | (Beishova, et al., 2018) |
|
Limousine (n = 100) |
0.642 | 0.358 | 0.461 | 0.363 | 0.176 | Poland | (Dybus, et al., 2002) |
Table 4: Frequencies of alleles and genotypes of bIGF-1-SnaBI polymorphism in the Holstein cattle bred in Kazakhstan.
| Allele | Observed allele frequencies | Relative allele frequencies | Genotype | Number of genotypes | Frequencies of genotypes |
|
bIGF-1-SnaBIА |
116 | 0.58 ± 0.005 |
bIGF-1-SnaBIАА |
18 | 0.18 |
|
bIGF-1-SnaBIB |
84 | 0.42 ± 0.005 |
bIGF-1-SnaBIАВ |
48 | 0.48 |
|
bIGF-1-SnaBIВВ |
34 | 0.34 | |||
| Total | 100 | 1 |
(Lucy et al., 1993; Sadeghi et al., 2008; Hartatik et al., 2015; Dolmatova and Ilyasov, 2011; Beishova et al., 2018; Fedota et al., 2016; Dybus et al., 2002) The obtained results correspond to the published data, and are within the frequency limits for the alleles obtained by various authors for both milk and beef breeds.
Table 5: Distribution of the relative frequencies of the alleles of bIGF-1-SnaBI polymorphism in various populations of Holstein cattle.
| Breed | Relative allele frequencies | Frequencies of genotypes | Country | Author, reference | |||
|
bIGF-1-SnaBIА |
bIGF-1- SnaBIB |
bIGF-1-SnaBIАА |
bIGF-1-SnaBIАВ |
bIGF-1-SnaBIВВ |
|||
| Dairy cattle breeds | |||||||
|
Holstein-Friesian (n = 42) |
0.63 | 0.37 | 0.33 | 0.59 | 0.07 | Uruguay | (Nicolini et al., 2013) |
|
Holstein-Friesian (n = 28) |
0.52 | 0.48 | 0.28 | 0.46 | 0.25 | New Zealand | (Nicolini et al., 2013) |
|
Holstein-Friesian (n = 662) |
0.52 | 0.48 | 0.29 | 0.47 | 0.24 | Poland | (Siadkowska et al., 2006) |
|
Holstein (n = 282) |
0.438 | 0.562 | 0.159 | 0.557 | 0.284 | Iran | (Mehmanna et al., 2010) |
| Beef cattle breeds | |||||||
| Korean | 0.72 | 0.28 | 0.586 | 0.264 | 0.150 | Korea | (Chung and Kim, 2005) |
|
Charolais (n = 68) |
0.46 | 0.54 | 0.208 | 0.500 | 0.292 | Mexico | (Reyna, et al., 2010) |
|
Canchim (n = 30) |
0.35 | 0.65 | 0.133 | 0.433 | 0.434 | Brazil | (Curi et al., 2005) |
|
Charolais (n = 43) |
0.26 | 0.74 | 0.070 | 0.372 | 0.558 | Mexico | (Reyna, et al., 2010) |
|
Beefmaster (n = 25) |
0.03 | 0.97 | 0 | 0.067 | 0.933 | Mexico | (Reyna, et al., 2010) |
|
Nelore (n = 79) |
0 | 1 | 0 | 0 | 1 | Brazil | (Curi et al., 2005) |
Table 6: Distribution of the frequencies of the genotypes of the somatotropin cascade polymorphic genes in the populations of Holstein cattle bred in Kazakhstan.
| Polymorphism | Genotype | n observed | n expected |
χ2 |
|
bGH-AluI n = 100 |
bGH-AluIVV |
4 | 3 | 0.15 |
|
bGH-AluILV |
29 | 30 | ||
|
bGH-AluILL |
67 | 66 | ||
|
bIGF-1-SnaBI n = 100 |
bIGF-1-SnaBIВВ |
34 | 34 | 0.02 |
|
bIGF-1-SnaBIАВ |
48 | 49 | ||
|
bIGF-1-SnaBIАА |
18 | 18 |
As a result of the DNA diagnostics of the Kazakhstan population of Holstein cows by the polymorphic locus of bIGF-1gene, it has been found that out of 100 animals, 18 had bIGF-1-SnaBIAA genotype, 48 cows had bIGF-1-SnaBIAB genotype, and 34 cows had bIGF-1-SnaBIBB genotype. With that, the frequency of bIGF-1-SnaBIA allele was 0.58, and that of bIGF-1-SnaBI allele–0.42 (Table 4).
A number of studies have been devoted to defining the polymorphism of the insulin-like growth factor-1 gene in cattle (Table 5) (Mehmannavaz et al., 2010; Siadkowska et al., 2006; Nicolini et al., 2013; Chung and Kim, 2005; Reyna, et al., 2010; Curi et al., 2005).
Studying the SnaBI- polymorphism of bIGF-1 gene showed that in Holstein and Holstein-Frisian cattle breeds, the frequency of bIGF-1-SnaBIA allele was 0.438 – 0.630, and the frequency of bIGF-1-SnaBIB allele– 0.370 – 0.562, respectively. In the beef breeds (Korean, Charolais, Canchim, Beefmaster, Nelore), the frequency of bIGF-1-SnaBIA allele was in the range between 0 and 0.72, while the frequency of bIGF-1-SnaBIB allele–between 0.28 and 1. In the study of the authors, the frequency of bIGF-1-SnaBIA and bIGF-1-SnaBIB alleles was medium, and amounted to 0.58 and 0.42, respectively.
The authors have also analyzed the conformity of the observed frequencies of the genotype to the theoretically expected equilibrium distribution in accordance with the law of Hardy-Weinberg (Table 6). The significance of the observed deviations was assessed using criterion χ2.
Table 6 shows that for Holstein cattle, for bGH-AluI and bIGF-1-SnaBI polymorphisms, the correspondence of the observed frequencies of the genotype to the ones theoretically expected is observed. This is evidence of the fact that the population of Holstein cows bred in Kazakhstan is genetically stable, and no natural and artificial selection is observed in it. Next, the authors plan to study the polymorphism of cattle LTF and MBL1 genes, and their association with economically useful traits.
CONCLUSION
Thus, the authors have found the following. The rare to common alleles ratio for bGH-AluI polymorphism coincides with the populations of Holstein cattle bred in various countries. The work has established that, like in other works, the rare one is bGH-AluIV allele. This observation suggests that allele of bGH-AluIL may provide selective advantages on the background of feeding peculiarities or climatic conditions. Allele frequencies of bIGF-1-SnaBI polymorphism, which were found in the population of Holstein cows bred in Kazakhstan, correlate with those established by other researchers. Unlike in the dairy ones, in the meat breeds, the frequency of bIGF-1-SnaBIB allele is higher; it is possible since along with the development of meat qualities, animals of this breed with excellent genotype were selected.
The results of analyzing the distribution of genotypes show that the observed genotype frequencies in the local Kazakhstan population of Holstein cows by bGH-AluI and bIGF-1-SnaBI polymorphisms coincide with those expected according to the law of Hardy-Weinberg, which indicates the absence of artificial selection in the population.
ACKNOWLEDGEMENT
The work has been performed in the framework of the project for grant funding of the Ministry of Education and Science of the Republic of Kazakhstan for years 2018–2020 “Development and implementation of a comprehensive program to improve productive longevity of high yielding cows of local selection” (State Registration No. 0118RK00398).
Authors Contribution
All authors contributed equally.
Conflict of interest
The authors declare no conflicts of interest.
References






