Advances in Animal and Veterinary Sciences
Research Article
Phylogenetic Analysis of Bovine Herpesvirus Isolate of India
Kota Sri Naga Leela Surendra1, Samir Kumar Rana1, Bhaskaran Mohana Subramanian2, Rachamreddy Venkata Chandrasekhar Reddy1, Girish Kumar Sharma3, Villuppananoor Alwar Srinivasan1*
1NDDB R&D Laboratory, National Dairy Development Board, Gachibowli, Hyderabad, India; 2Translational Research Platform for Veterinary Biologicals, Chennai, India; 3National Dairy Development Board, Animal Health, Anand, India.
Abstract | Bovine herpesvirus-1 (BHV-1) causes infectious bovine rhinotracheitis (IBR) and infectious pustular vulvovaginitis / balanoposthitis (IPV/IPB) in cattle and buffaloes. BHV-1 is closely related to bovine herpesvirus-5 (BHV-5), which has been recovered from both genital and respiratory tracts of cattle and buffaloes. Perusal of literature reveals paucity of information on genetic characteristics of BHV subtypes circulating in India. In the present study, 25 isolates originated from five different states of India viz. Gujarat, Uttar Pradesh, Maharashtra, Karantaka and Andhra Pradesh, during the period 1983-2010, were genetically characterized by restriction endonuclease analysis (REA) and partial sequencing of unique long (UL27, UL44) and unique short regions (US1.67) of the viral genome. The REA patterns (Hind III) of these isolates indicated that they were indistinguishable from BHV-1.1 subtype. The phylogenetic analysis based on nucleotide sequences of the unique long (UL) and unique short (US) regions revealed the high sequence homology (>99%) within the isolates and their close relatedness to the BHV-1.1 subtype. The present study established the prevalence of BHV-1.1 as the predominant circulating subtype of BHV in India. The current study also confirmed that the genomic fingerprinting based on HindIII cleavage, direct sequencing of gC (UL44) and gB (UL27) gene-derived PCR amplicons were useful tools for genetic characterization of BHV strains.
Keywords | Bovine herpesvirus, Restriction endonuclease analysis, Phylogenetic analysis, Glycoprotein, BEAST, India
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | June 11, 2015; Revised | June 23, 2015; Accepted | June 25, 2015; Published | July 01, 2015
*Correspondence | Villuppananoor Alwar Srinivasan, National Dairy Development Board, Animal Health, Hyderabad, India; Email: [email protected]
Citation | Surendra KSNL, Rana SK, Subramanian BM, Reddy RVC, Sharma GK, Srinivasan VA (2015). Phylogenetic analysis of bovine herpesvirus isolate of India. Adv. Anim. Vet. Sci. 3(8): 451-460.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.8.451.460
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Surendra et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Bovine herpesvirus-1 (BHV-1) causes Infectious Bovine Rhinotracheitis (IBR) and Infectious Pustular Vulvovaginitis/Balanoposthitis (IPV/IPB) in cattle and buffaloes. BHV-1 is a double stranded DNA virus of the genus Varicellovirus, subfamily Alphaherpesvirinae and family Herpesviridae. The disease causes high morbidity and low mortality and has been associated with a spectrum of other clinical manifestations such as encephalitis, conjunctivitis, enteritis, abortions and reduction in milk yield (Gibbs and Rweyemamu 1977; Hage et al., 1998; Nandi et al., 2009). Three different genotypes of BHV-1 (BHV-1.1, BHV-1.2a and BHV-1.2b) had been reported so far based on the genomic characteristics and antigenic properties of the virus (Metzler et al., 1985; D’Arce et al., 2002; Souza et al., 2002; Muylkens et al., 2007). A closely related virus, Bovine herpesvirus-5 (BHV-5), was also occasionally been recovered from both genital and respiratory tracts of cattle and the BHV-5 infection causes bovine encephalitis or meningoencephalitis in cattle and buffaloes (Bratanich et al., 1991; Mahony, 2010). Recent reports confirm the occasional association of BHV-1 with encephalitis (Roels et al., 2000). However, the mortality rate in younger calves due to BHV-5 infection is far higher (~100 %) than the mortality due to BHV-1 infection (Studdert, 1989). Genomes of BHV-1 and BHV-5 share high sequence homology (~85%) and these viruses display extensive serological cross-reactivity in most of the serological tests employed (Roehe et al., 1997; Campos et al., 2009).
Subsequent to the report of BHV-1 in Indian cattle during1976 (Mehrotra et al., 1976), several authors reported varying sero-prevalence rates ranging from 50.9 to 60.46% and 75 to 81.0% in Indian cattle and buffalo population, respectively (Renukaradhya et al., 1996; Renukaradhya et al., 2002; Malmurugan et al., 2004; Trangadia et al., 2010). More than 45 percent of apparently healthy breeding bulls were found to be seropositive for IBR/IPV and 20 per cent of IBR/IPV seropositive bulls showed intermittent shedding of virus in semen (Deka et al., 2005; Rana et al., 2011). Isolation of BHV-1 from Indian cattle was reported earlier, albeit with a very few virus isolates (Mehrotra et al., 1976; Chandranaik et al., 2010; Saha et al., 2010; Ravishankar et al., 2012). Prevalent BHV-1 types and subtypes can only be identified based on the established methods of genetic characterisation (Metzler et al., 1985; D’Acre et al., 2002) and prevalence of BHV-5 can also be ruled out/established by these methods. In the present study, genetic characterisation of the Indian isolates of BHV, which were obtained from various geographical regions of India during 1983-2010, was performed by restriction endonuclease fingerprinting of the whole viral genome and partial nucleotide sequence analysis.
Materials and Methods
Cells and Semen Samples
Monolayers of Madin-Darby bovine kidney cells (MDBK) for virus isolation were obtained from cell culture laboratory, R&D, National Dairy Development Board, India. Extended frozen semen of cattle and buffalo were procured from various semen banks in India.
Virus Isolation in Cell Culture
Virus isolation was carried out from extended frozen semen / genital and nasal swabs collected from IBR seropositive cattle as per the procedure of OIE manual (2010). Briefly, 1ml of extended semen was diluted with foetal bovine sera (Hyclone-FBS, New Zealand), inoculated onto MDBK cell monolayer and incubated at 37ºC. The monolayer was observed periodically under a microscope for the appearance of cytopathic effect (CPE) until day seven post inoculation and three such consecutive passages were carried out. The BHV-1 isolates were further confirmed by virus neutralization test (VNT) and PCR. The virus neutralization test was performed using BHV-1 positive serum following the procedure described in the OIE manual (OIE manual, 2010). Total DNA was extracted from the tissue culture supernatant using a QIAamp DNA Blood mini kit (Qiagen, Germany) and the DNA were subjected to PCR using gB gene and gE gene specific primers and the details of primers were depicted in Table 1 (Abril et al., 2004; Monika et al., 1999). PCR was carried out in a total of 50µl reaction volume containing: Taq PCR buffer with 1.5mM MgCl2, 0.2mM of dNTPs each, 30pM (0.6 µM) of each primer, 1.5 units of Taq DNA polymerase enzyme (Qiagen, Germany ), 1X Q- solution (Qiagen, Germany) and 50ng of the DNA sample.
Virus Purification and DNA Extraction
MDBK cell monolayer was infected with the virus isolates for amplification. The virus infected cells were incubated at 37ºC until 90% CPE was observed and the cell culture supernatant was used for virus purification. The cell debris was removed from the culture supernatant by centrifugation at 3000 g for 15 minutes and the virus was pelleted by centrifuging the clarified culture supernatant at 53,000g using P28S2 rotor for 2.5 hours in a Hitachi preparative ultracentrifuge. The pellet was resuspended in 500μl Tris-NaCl EDTA buffer (0.1M NaCl, 10mM Tris-HCL, 1mM EDTA; pH 8.0). The virus was purified using sucrose density gradient centrifugation. The resuspended virus pellet was layered over a step gradient with 30%, 40% and 60% sucrose and centrifuged at 120,000g for 2.5hrs in a Beckman preparative ultracentrifuge using SW40Ti swing out rotor. The purified virus was collected from the interface of 40% and 60% sucrose. DNA was extracted from the purified virus suspension using a QIAamp DNA Blood mini kit (Qiagen, Germany) as per manufacturer’s instruction. DNA concentration was determined using BioPhotometer (Eppendorf, Germany). The purity and integrity of the extracted DNA was checked by electrophoresis in 0.6% agarose gel.
Restriction Enzyme (RE) Analysis
The viral DNA was digested using restriction enzymes HindIII according to the manufacturer’s instructions (New England Biolabs Inc., USA). The digestion reaction was performed in a 30µl volume consisting of 1x reaction buffer, 3µg purified viral DNA and 100 units of HindIII at 37°C for 16 hrs. The digested products were resolved in a 0.6% agarose gel which was run using 1x Tris borate EDTA buffer (89mM boric acid, 2mM EDTA, 89mM Tris-HCL; pH 8.0) at 30V for 20 hours. HindIII-digested lambda phage DNA was used as a molecular weight marker. Similarly the viral DNA was digested with BstEII, so as to attain the REA profile of the viral genomes. Based on the electrophoretic profile, all the isolates were classified according to the system proposed by Metzler et al. (1985).
Partial Nucleotide Sequencing and Phylogenetic Analysis
Partial sequence analysis of unique short region (US1.67, Wang et al., 2006) and unique long region (UL27 & UL44, Esteves et al., 2008; Momtaz, 2009) was performed. The regions of partial US 1.67, UL27 (glycoprotein B; gB) and UL 44 (glycoprotein C; gC) were amplified by PCR with resultant product sizes of 438 bp, 535 bp and 666 bp respectively (Table 1). The PCR products were gel purified (QIAquick gel purification kit, Qiagen, Germany)
Table 1: Details of primer sequences used for PCR and partial nucleotide sequencing
|
Target gene |
Primer Sequence (5' - 3') |
Details of Nucleotide position in Genome |
GenBank® Accession number |
Amplicon size (Base pair-bp) |
|
Polymerase chain reaction |
||||
|
gB |
gB-F:TGT-GGA-CCT-AAA-CCT-CAC-GGT |
57499–57519 |
AJ004801 |
97 |
|
gB-R: GTA-GTC-GAG-CAG-ACC-CGT-GTC |
57595–57575 |
|||
|
gE |
gE F: CTT-CGG-TCG-ACA-CGG-TCT-T |
501–520 |
U06934 |
265 |
|
gE R - CTT-TGT-CGC-CCG-TTG-AGT-CG |
746-765 |
|||
|
Phylogenetic analysis |
||||
|
US1.67 |
F- AGC-GGG-GCC-TCG-TCC-TCG-TAG-CAC |
114552 -114575 |
AJ004801 |
438 |
|
R- CAG-CGC-CGG-CGT-TTG-GTC-ATT-TG |
114967 -114989 |
|||
|
UL44 (gC) |
F- CGG-CCA-CGA-CGC-TGA-CGA |
16762 -16779 |
AJ004801 |
575 |
|
R- CGC-CGC-CGA-GTA-CTA-CCC |
17319 -17336 |
|||
|
UL27(gB) |
F –GTA-CAC-GTT-CAA-GGC-CTA-CA |
55896 – 55915 |
AJ004801 |
666 |
|
R-TCG-TCT-CGC-AGC-ATT-TC |
56546 -56561 |
|||
Table 2: Details of BHV-1 isolates from different states of India
|
Sr. No |
Isolate ID |
State/Country |
Origin |
|
1 |
GUK 1/83 |
Maharashtra/India |
Semen |
|
2 |
MAA 2/01 |
Maharashtra/India |
Semen |
|
3 |
MAA 3/01 |
Maharashtra/India |
Semen |
|
4 |
MAA 4/01 |
Maharashtra/India |
Vaginal/Genital swab |
|
5 |
MAA 5/01 |
Maharashtra/India |
Vaginal/Genital swab |
|
6 |
MAA 6/01 |
Maharashtra/India |
Semen |
|
7 |
KAA 01 |
Karnataka/India |
Semen |
|
8 |
GUK 124/07 |
Gujarat/India |
Semen |
|
9 |
GUK 144/07 |
Gujarat/India |
Semen |
|
10 |
GUK 145/07 |
Gujarat/India |
Semen |
|
11 |
GUK 55/07 |
Gujarat/India |
Semen |
|
12 |
GUK 56/07 |
Gujarat/India |
Semen |
|
13 |
GUK 57/07 |
Gujarat/India |
Semen |
|
14 |
GUK 99/07 |
Gujarat/India |
Semen |
|
15 |
UPR 27/07 |
Uttar Pradesh/India |
Semen |
|
16 |
UPR 28/07 |
Uttar Pradesh/India |
Semen |
|
17 |
UPR 29/07 |
Uttar Pradesh/India |
Semen |
|
18 |
UPR 31/07 |
Uttar Pradesh/India |
Semen |
|
19 |
UPR 33/07 |
Uttar Pradesh/India |
Semen |
|
20 |
UPR 34/07 |
Uttar Pradesh/India |
Semen |
|
21 |
UPR 38/07 |
Uttar Pradesh/India |
Semen |
|
22 |
UPR 39/07 |
Uttar Pradesh/India |
Semen |
|
23 |
UPR 40/07 |
Uttar Pradesh/India |
Semen |
|
24 |
APH 01/10 |
Andhra Pradesh/India |
Nasal Swab |
|
25 |
APH 02/10 |
Andhra Pradesh/India |
Vaginal/Genital Swab |
and subjected to cycle sequencing. Cycle sequencing was performed using the BigDye® Terminator v3. 1 Cycle Sequencing Kit (Applied Biosystems, Part No- 4337455). The cycle sequencing products were purified and loaded on ABI PRISM® 3130xl genetic analyzer (Applied Biosystems®, USA) for capillary gel electrophoresis through the performance optimized polymer (POP-7TM). The raw data were collected using data collection software v3. 0 and nucleotide sequences were analyzed using sequencing analysis software v5.2. Accuracy of the DNA sequences was checked using SeqScape® v2.5 (Applied Biosystems) and GeneDoc (Nicholas et al., 1997).
Apart from the sequences of the twenty five isolates of the present study (Table 2), sequences of representative bovine herpesvirus isolates and related herpesvirus were retrieved from GenBank and were included in the phylogenetic analysis (Table 3). The sequences were aligned using ClustalW in MEGA version 6.06 (Tamura et al., 2013). Following multiple alignment for these sequenced regions, the Bayesian Information Criterion (BIC), maximum likelihood values and Akaike Information Criterion corrected (AICc) scores were also determined for the maximum likelihood fits based on the data specific model to generate the phylogenetic tree for US 1.67, UL27 and UL44. The ML tree topology was evaluated using both neighbour-joining (NJ) and ML methods with 1,000 and 500 bootstrap replicates respectively. Details of all the sequences, which were used in the study, were provided in Table 2. The nucleotide sequences of regions targeted in the present study were compared with the corresponding sequences of BHV-1 and BHV-5 retrieved from GenBank.
Table 3: Information on Bovine herpesvirus and related virus isolates used in the present study for partial nucleotide and phylogenetic analysis
|
Sr. No |
Isolate ID |
Country |
Year of Isolation |
Accession Numbers |
|
1 |
BHV-1.1/MN1 |
USA |
2004 |
KJ652513.1*, KJ652521.1ǂ |
|
2 |
BHV-1.1/MN2 |
USA |
2008 |
KJ652514.1*, KJ652522.1ǂ |
|
3 |
BHV-1.1/MN3 |
USA |
2009 |
KJ652515.1*, KJ652523.1ǂ |
|
4 |
BHV-1.1/MN4 |
USA |
2010 |
KJ652516.1* |
|
5 |
BHV-1.1/MN5 |
USA |
2010 |
KJ652517.1* |
|
6 |
BHV-1.1/MN6 |
USA |
2012 |
KJ652518.1* |
|
7 |
BHV1.1/Abu-Hammad/1 |
Egypt |
2013 |
KJ652519.1*, KJ652527.1ǂ |
|
8 |
BHV1.1/Abu-Hammad/2 |
Egypt |
2013 |
KJ652520.1*, KJ652528.1ǂ |
|
9 |
BHV-1.1/COLORADO |
USA, Philadelphia |
1988 |
M21474.1*, |
|
10 |
BHV-1/NM06 |
China |
2006 |
JN787952.1*, JN787953.1ǂ |
|
11 |
BHV-1.1/COOPER |
USA |
1956 |
AJ004801.1*♦ |
|
12 |
BHV-1.1 NVSL |
USA |
- |
JX898220.1♦ |
|
13 |
BHV-1.2 ST |
- |
- |
Wang et al., 2006 |
|
14 |
BHV-1 PDADMAS |
India |
2008 |
EU523747♦ |
|
15 |
BHV-1.2b/SP1777 |
USA |
2009 |
KM258883.1*ǂ♦ |
|
16 |
BHV-1.2b/SM023 |
USA: Texas |
1986 |
KM258882.1*ǂ♦ |
|
17 |
BHV-1.2b/B589 |
Australia |
2001 |
KM258881.1*ǂ♦ |
|
18 |
BHV-1.2b/K22 |
USA: New York |
1958 |
KM258880.1*ǂ♦ |
|
19 |
BHV-5/SV507/99 |
Brazil: southern |
1999 |
AY261359.1*, AY261359.2ǂ |
|
20 |
BHV-5/EVI-88 |
Brazil |
1988 |
AY330350.1*, DQ173720.1ǂ |
|
21 |
BuHV-1/B6" |
Switzerland |
2001 |
AF359760.1* |
|
22 |
CHV-1/Banffshire 82 |
Switzerland |
1998 |
AF078729.2* |
|
23 |
RHV-1/Salla 82 |
Switzerland |
1998 |
AF078727.2* |
|
24 |
BHV-1.1/UY1999 |
Uruguay |
1999 |
DQ173735.1ǂ |
|
25 |
BHV-1.1/ LAM |
Netherlands |
- |
DQ173724.1ǂ |
|
26 |
BHV-1.1/COOPER |
USA |
1956 |
DQ173733.1ǂ♦ |
|
27 |
BHV-1/Cro08 |
Croatia |
2008 |
GQ169130.1ǂ |
|
28 |
BHV-1/TRP1 |
Brazil |
1998 |
KJ634157.1ǂ |
|
29 |
BHV-1/Iso215/75 |
India |
1975 |
JX127198.1ǂ |
|
30 |
BHV-1/Iso216/75 |
India |
1975 |
JX127201.1ǂ |
|
31 |
BHV-1/7E |
India |
2012 |
JX127195.1ǂ |
|
32 |
BHV-1/NM06 |
China |
2006 |
JN787953.1ǂ |
|
33 |
BHV-1.2/SV265 |
Brazil |
2005 |
DQ173718.1ǂ |
|
34 |
BHV-1.2/UY2002 |
Uruguay |
2002 |
DQ173731.1ǂ |
|
35 |
BHV-1.2/UY2004 |
Uruguay |
2004 |
DQ173732.1ǂ |
|
36 |
BHV-5/ISO87 |
Brazil |
1987 |
DQ173725.1ǂ |
|
37 |
BHV-5/TX89 |
USA |
1989 |
U35883.1ǂ |
|
38 |
BHV-5/EVI340 |
Brazil |
1996 |
DQ173734.1ǂ |
|
39 |
BHV-5/SV136 |
Brazil |
1988 |
DQ173722.1ǂ |
|
40 |
BHV-5/EVI88 |
Brazil |
1995 |
DQ173720.1ǂ |
|
41 |
CpHV-1/Ba-1 |
Italy |
1996 |
AY821804.1ǂ |
|
42 |
CpHV-1 |
Switzerland |
1995 |
Z49225.1 ǂ |
|
43 |
CvHV-1 |
Canada |
2005 |
DQ333390.1ǂ |
|
44 |
CvHV-2 |
Canada |
2005 |
DQ333391.1 ǂ |
|
45 |
BuHV-1/izsm.13 |
Italy |
2012 |
KF679678.1ǂ |
|
46 |
BHV-1/02/1838 NZ |
New Zealand |
- |
Wang et al., 2006♦ |
|
47 |
BHV-1/01/152_NZ |
New Zealand |
- |
Wang et al., 2006♦ |
|
48 |
BHV-1/00/2834_NZ |
New Zealand |
- |
Wang et al., 2006♦ |
* - GenBank Sequence considered for drawing NJ Tree based on UL27; ǂ - GenBank Sequence considered for drawing NJ Tree based on UL44; ♦ - GenBank Sequence considered for drawing NJ Tree based on US 1.67
Evolutionary Analysis
Evolutionary analysis was performed using BEAST program (BEAST software package, v1.8.0; Drummond et al., 2012) for the partial gB gene sequences (Table 3). Bayesian maximum clade credibility phylogenetic tree with Bayesian Markov Chain Monte Carlo (MCMC) analysis was created. The input data for the Beast analysis was obtained using BEAUti software v1.8.0 and year of the virus isolation was tip dated. The GTR+I+G nucleotide substitution model was determined as the best fit based on the Akaike Information Criterion (AIC) scores and an uncorrelated lognormal relaxed clock model was chosen. The MCMC chains were run for a chain length of 2 x108 and sampled at every thousand generations. The nucleotide substitution rate (substitutions/site/year) and the time to Most Recent Common Ancestor (tMRCA) values were obtained from the Tracer, v1.5. The posterior tree distributions were summarized using Tree Annotator with the exclusion of the initial ten percent of trees and visualised in FigTree v1.3.1.
Results
Virus Isolation in Cell Culture
Monolayers of MDBK cells were inoculated with semen samples or genital/nasal swab samples for isolation of the virus (OIE Manual, 2008). In 25 cell monolayers to which samples were added, microscopic examination revealed the presence of grape-like clustered and rounded call. Presence of giant cells is considered as characteristic cytopathic effect of BHV infection in cell culture. However, the numbers of days for CPE differed for each sample. Nine samples showed characteristic CPE in the first passage after 4 days to 5 days of inoculation. Whereas in similar effect was observed (16 samples) after 24 hours (1 day) in the second passage. Presence of CPE in any of the three passages was considered as positive for virus isolation. A total of 183frozen semen samples, 50 genital swabs, 25 nasal swabs were processed for virus isolation and CPE was observed in 21, 3 and 1 samples, respectively (Table 2).
Virus Neutralisation Test (VNT) and PCR
Virus neutralization test (VNT) and PCR were used to confirm the identity of all the 25 isolates which showed CPE in cell culture. Those samples, which showed more than two log reduction in virus infectivity in the presence of known positive serum, were considered as VNT positive. All the 25 isolates turned positive for VNT. PCR using gB and gE gene specific primers produced PCR amplicons of 97 bp and 265 bp sizes, respectively. The VNT and PCR results confirmed the cytopathogenic agent as bovine herpesvirus.
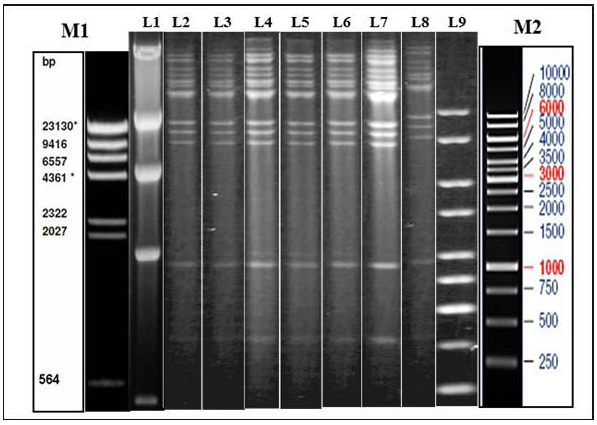
Figure 1: Illustration of Restriction endonuclease (RE) analysis profile of Indian isolates of the present study
a) Agarose gel showing the electrophoretic profile of representative viral genome digested with Hind III. Lane 2-8 represents Indian isolates - GUK 01/83, GUK 56/07, KAA 01, MAA 4/01, UPR 29/07, UPR 38/07, APH 01/10 respectively. M1 & L1 – Lambda Hind III Digest; M2 & L9 – Fermentas 1 Kb Gene ruler
Restriction Enzyme (RE) Analysis
Genomic DNA was extracted from sucrose density gradient purified virus isolates. The viral DNA was subjected to RE using HindIII and BstEII restriction enzymes separately. The electrophoretic profiles of the HindIII digested viral DNA were similar for all the virus isolates which originated from Gujarat, Uttar Pradesh, Maharashtra and Andhra Pradesh (Figure 1). The RE pattern of these isolates were compared with that of patterns of BHV-1.1, BHV1.2 and BHV-5 (Meltzer et al., 1985) and the RE profiles of the Indian isolates had a close similarity with the BHV-1.1 subtype. RE profile of BstEII displayed some heterogeneity among the Indian isolates. Few of the isolates (UPR 40/07, GUK 57/07 and GUK 145/07) showed an additional band at 2.5Kb; all isolates from Uttar Pradesh and Gujarat displayed two distinct bands in the 4.0 Kb to 9.4Kb region; isolates from Maharashtra showed only one band in this region (data not shown). Though, there were some variation between the isolates in the REA profile of BstEII, the electrophoretic profile of these isolates was distinctly different from the REA pattern of BHV-5.
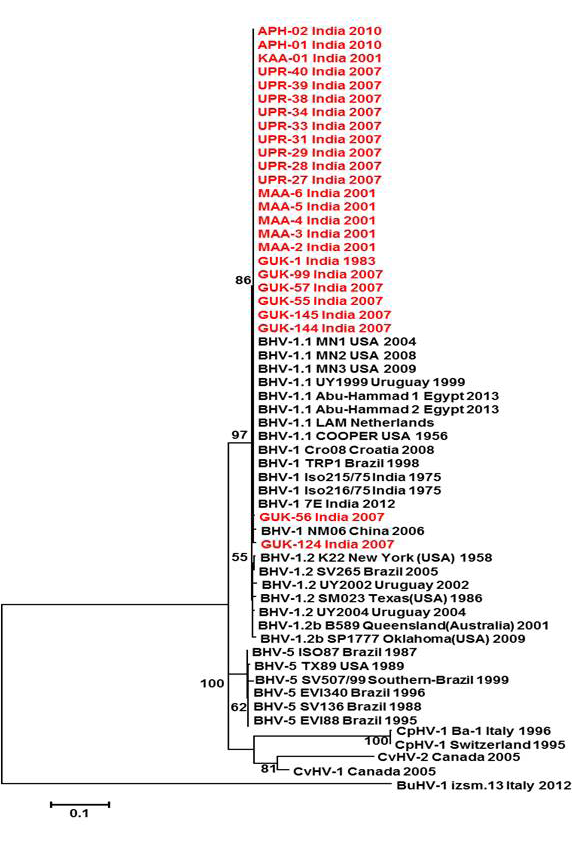
Figure 2: Neighbour- joining tree generated by 454 bp of glycoprotein C (UL 44) region showing the genetic relationship of the Indian isolates with published bovine herpesvirus and related herpesviruses. The percentage of bootstrap values given to the left of main branch
Partial Nucleotide Sequencing and Phylogenetic Analysis
Sequencing and Sequence Homology
Partial sequences of US1.67, UL27 (gB gene) and UL44 (gC gene) of all the 25 Indian isolates were generated by cycle sequencing. Partial sequences with the read lengths of 304bp (US1.67), 624bp (UL27) and 454bp (UL44), were used for sequence analysis. These sequences were compared with the corresponding sequences of GenBank (Table 2). Nucleotide sequences of these Indian viruses were submitted to GenBank with Accession nos. FJ969564 to FJ969585 and KF734594 to KF734647 (Table 2 and Table 3).
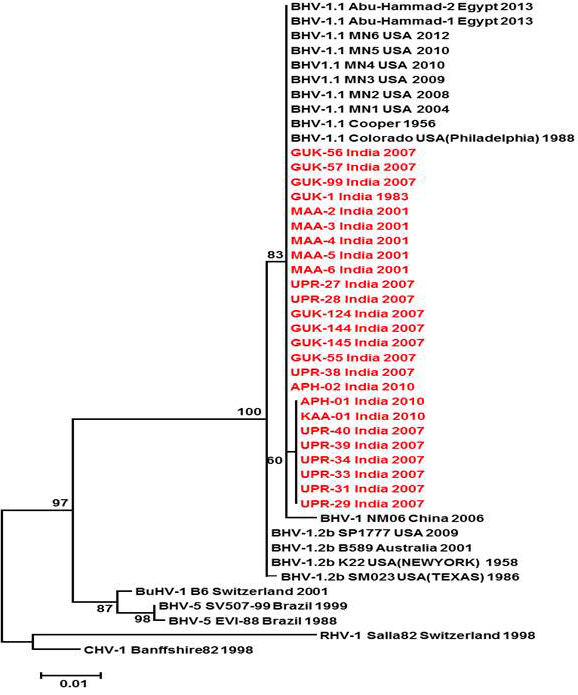
Figure 3: Neighbour- joining tree generated by 624 bp of glycoprotein B (UL 27) region showing the genetic relationship of the Indian isolates with published bovine herpesvirus and related herpesviruses. The percentage of bootstrap values given to the left of main branch
Partial US1.67 sequences of the present study exhibited 99.01 to 99.34 % similarity among them and the sequence identity was ~99.34 with the available BHV-1.1 (BHV-1.1 Cooper and Jura 1.1), followed by 98.99% homology with BHV-1.2 (ST Strain). Similarly, the partial UL27 sequences showed 100% homology with many published BHV-1.1 sequences. Variable levels of identity were recorded with BHV-1.2 strains (~99.66%), BHV-5 (~95.14 – 95.31%), BuHV-1(~95.64), CHV-1 and 2 strains (94.29%) and CpHV-1(~87.44). The partial UL44 sequences of the Indian isolates showed high similarity with BHV-1.1 (99.11 – 100.00%) followed by BHV-1.2 (98.45-99.34), BHV-5(89.6-92.48%), CvHV-1 and 2 (79.87-88.05%) and CpHV-1(78.98-79.42%). The sequence (UL44) identity among isolates of the present study was greater than 99.33%.
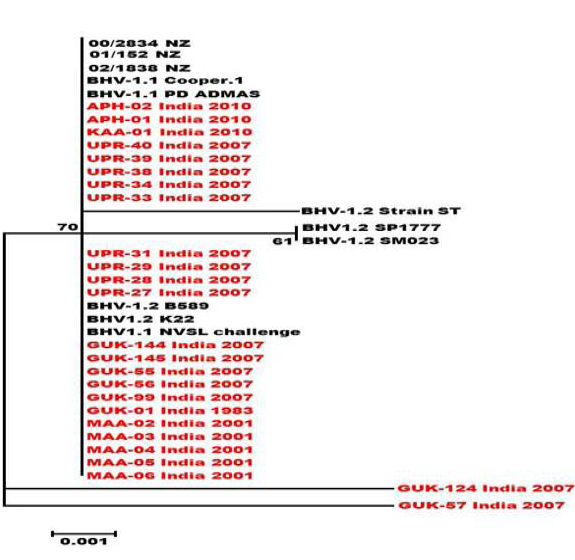
Figure 4: Neighbour- joining tree generated by 304 bp of US 1.67 region showing the genetic relationship of the Indian isolates with related herpesviruses. The percentage of bootstrap values given to the left of main branch
Phylogenetic Analysis
Neighbour joining tree was constructed for the partial sequences of UL44, UL27 and US1.67 regions (Figure 2, Figure 3 and Figure 4). The maximum clade credibility tree using the partial nucleotide sequences of gC and gB gene showed similar topology of clustering for BHV-1, BHV-5 and other related herpesvirus as described earlier (Esteves et al., 2008) (Figure 2 and Figure 3). The isolates of the present study were found clustering along with reference BHV-1 strains in all the three Phylogenetic trees. These isolates formed a distinctly separate lineage in the NJ trees compared to BHV-5 and other related alphaherpesvirus (CvHV-1, CpHV-1, BuHV-1, CHV-1 and RHV-1) (Figure 2 and Figure 3). These results clearly indicate that the Indian BHV belongs to BHV-1 group and there is no evidence for the presence of BHV-5. In the partial gC and gB gene sequence based NJ trees, the isolates of the present study and the BHV-1.1 reference isolates were in a distinct cluster compared to BHV-1.2 subtype isolates.
The maximum clade credibility tree was drawn using BEAST program for the partial gB gene sequences (Figure 5). General topology of the evolutionary tree was similar to the partial gB gene sequence based NJ tree. Our inferred substitution rate for the partial gB gene sequences was 1.23 X 10-3 (95% HPD; 1.24 X 10-3 to 1.222 x 10-3). There was not any distinct evolutionary pattern for the Indian isolates of BHV 1 (Figure 5). It was also evident that the sub types of BHV 1 (BHV 1.1 and BHV 1.2) were evolving independent of each other from a common ancestor.
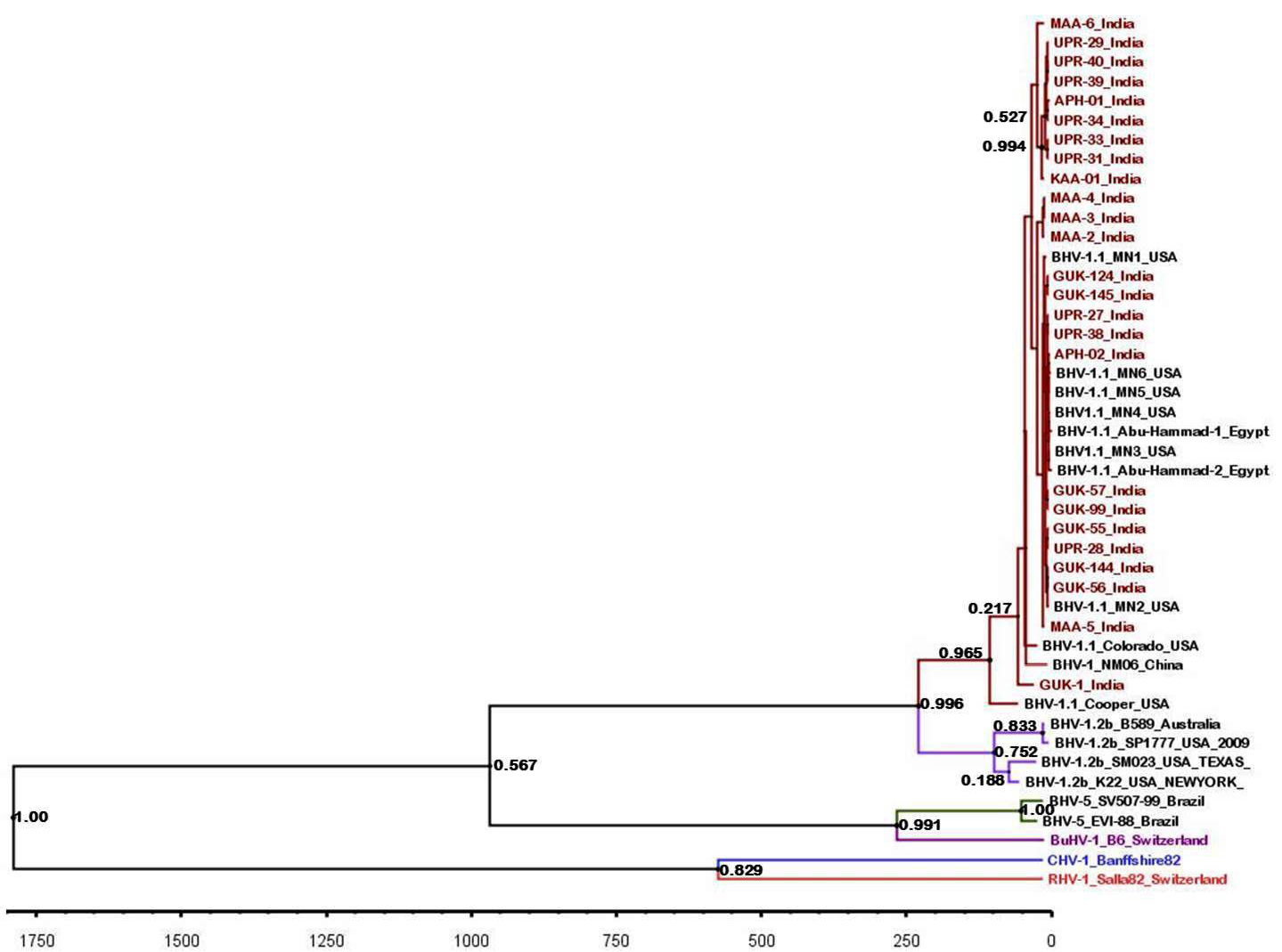
Figure 5: BEAST tree generated based on the BHV partial gB gene. The time scale is shown at the bottom
Discussion
Bovine herpesvirus causes respiratory, genital diseases and encephalitis in cattle with high morbidity and low mortality rates (Roels et al., 2000). The major mode of transmission in cattle and buffalo may occur through natural service or artificial insemination with infected semen (Van et al., 1995). In India, sero-epidemiological survey revealed 39.2 per cent of the cattle and buffaloes in organized herds were serologically positive for BHV (Nandi et al., 2010) while Trangadia et al. (2010) reported the BHV incidence as high as 60.84% in organized herds.
After an acute infection, the bovine herpesvirus enters sensory ganglia and establishes latency. The infected animal may become a carrier. Carrier animals tend to shed the virus intermittently (particularly under stress conditions) in nasal, ocular, vaginal secretions and in semen. It has been reported that approximately 20 percent of seropositive bulls shed virus intermittently in semen and ~2.2 percent of semen batches turned positive for BHV-1 (Rana et al., 2011). The increasing sero-prevalence, lack of IBR control programme and the presence of virus in semen are the foremost concerns in the Indian dairy industry as this disease is likely to have high economic impact. Though, the sero-prevalence of BHV has been reported in India more than a decade ago (Renukaradhya et al., 1996; Trangadia et al., 2010; Trangadia et al., 2012), reports on virus isolation and characterisation were scanty (Chandranaik et al., 2010; Saha et al., 2010; Ravishankar et al., 2012). The information on the prevalent types and subtypes of BHV in India is also lacking.
In the present study, bovine herpesvirus was isolated from extended frozen semen, genital and nasal swab samples collected from cattle and buffaloes using MDBK monolayer cells as prescribed in OIE Manual 2010. Total 25 virus isolates were obtained from the above specimens and 21, 3 and 1of those isolates were from frozen semen, genital and nasal swabs, respectively.
All these 25 isolates were typed according to the HindIII endonuclease digestion pattern as shown by Metzler et al., (1985). The size and pattern of HindIII fragments of all isolates in the present study were similar to the characteristics RE pattern of BHV-1.1 viral genomes (Metzler et al., 1985).The BstEII RE profiles of these isolates were distinctly different from that of BHV-5 (Metzler et al., 1985). On the basis of the results obtained in the current study, the Indian isolates could be classified as BHV-1.1 subtype. In India, the isolation of BHV-1.1 subtype from respiratory infection was also reported by Gupta et al. (1993) and Sreenivasa et al. (1996). Although, a recent investigation states the isolation of BHV-1.2 subtype from a nasal swab of cattle in India (Saha et al., 2010), none of the isolates of the present study could be classified as BHV-1.2.
Partial nucleotide sequence analysis of US1.67, UL27 and UL44 regions was performed to understand the genetic relatedness of the isolates with other strains circulating in the remaining parts of the world. The outcome of all the phylogenetic analysis is nearly the same and the analysis showed consistent grouping of Indian isolates along with bovine herpesvirus type 1.1 (BHV-1.1). BHV-1 and BHV-5 were clearly separated and grouped into different clusters in phylogenetic tree which was drawn based on partial nucleotide sequences of UL27 and UL44. The grouping of viruses according to their subtypes in Phylogenetic analysis based on the nucleotide sequencing of carboxy terminal of gC (UL44) were in harmony with findings of Esteves et al. (2008). However, a larger number of sequences from BHV-1.2 isolates are to be studied to ascertain whether the C-terminal partial gC gene based phylogenetic tree can be used to distinguish the BHV-1 subtypes.
Topology of partial gB gene sequence based evolutionary tree was similar to the one based on NJ tree. We could not identify any evolutionary lineage for the Indian isolates of BHV though IBR is a recently reported disease of Indian cattle. The BHV 1.1 and 1.2 sub-types were evolving independently of each other and might have got a common ancestor during the evolution.
The REA patterns of all Indian isolates (1983 to 2010) and phylogenetic analysis based on conserved unique short and unique long regions demonstrated that BHV 1.1 is the common subtype in India and this finding is also supported by results of Gupta et al. (1993) and Sreenivasa et al. (1996). None of the Indian isolates of the present study closely related to BHV-5 which was not reported in India so far. Further analysis of many isolates from various parts of India will establish the presence or absence of BHV-5.It is essential to establish the types and subtypes of circulating strains of bovine herpesviruses in India for formulating suitable control strategies and for the implementation of a suitable vaccination program.
In conclusion, the findings of the current study showed that genomic fingerprinting based on endonuclease HindIII cleavage, direct sequencing of partial gB and gC genes appear to be useful tools for genetic characterization of BHV strains. Phylogenetic analysis indicated the prevalence of BHV-1.1 types in India and the absence of BHV-5. The present study establishes the presence of BHV type which is responsible for abortion and other production losses in dairy industry in India. Therefore, an effective surveillance program is required for planning an appropriate control programme in India.
Acknowledgement
The authors are grateful to the National Diary Development Board (NDDB), Anand for providing the facilities to carry out this work conducted at the Research and Development (R&D) Laboratory, NDDB, Hyderabad.
The author K.S.N.L. Surendra express his gratitude to the Indian Immunologicals Limited, Hyderabad for providing him the opportunities to work on the topic of “Phylogenetic analysis of bovine herpesvirus isolates of India”, with respect to partial fulfilment for his Ph.D thesis.
Author’s Contribution
Kota Sri Naga Leela Surendra carried out laboratory work, conducted the experiments, analysed the data and prepared the manuscript. Samir Kumar Rana designed the study, isolated few viruses of present study, facilitated the study by arranging samples and finalised the manuscript. Bhaskaran Mohana Subramanian provided expert opinions on the design of work and assisted in analysis of nucleotide sequences and phylogenetic trees. Rachamreddy Venkata Chandrasekhar Reddy helped in nucleotide sequencing and annotation. Girish Kumar Sharma helped in collecting field specimens for virus isolation, provided the administrative support and facilitated financial support from the NDDB. All the authors are thankful to Villuppananoor Alwar Srinivasan for conceptualised the work from time to time, critically reviewed the manuscript and proving a meaningful conclusion from the data generated from the present study.
Conflict of Interest
The authors have no conflict of Interest.
References






