Advances in Animal and Veterinary Sciences
Research Article
Characterization of Rhodococcus equi Isolates from Foals with Respiratory Problems using a Multiplex PCR for the vap Genes
Sourabh Chhabra1, Sandip Kumar Khurana2*, Pradeep Kumar Kapoor1, Harisankar Singha2, Yarvendra Singh1, Richa Khirbat3
1Department of Veterinary Public Health and Epidemiology, LUVAS, Hisar; 2National Research Centre on Equines, Hisar; 3Department of Animal Biotechnology, LUVAS, Hisar, India.
Abstract | Rhodococcus equi is widely associated with subacute or chronic suppurative bronchopneumonia, ulcerative lymphangitis, lymphadenitis and enteritis in foals aged 1-4 months. Identification of the virulence plasmids associated with R. equi is of great importance in ascertaining pathogenicity in equines. Virulence factor is associated with presence of 85-90kb plasmid which encodes various virulence associated proteins (VAP -A,-B,-C,-D,-E,-F,-G,-H). VAP A is essential for virulence in foals. This study was aimed at the evaluation of molecular characteristics of R. equi isolates from foals with respiratory problems by multiplex PCR assay that amplifies the vap gene family (vap-A,-B,-C,-D,-E,-F,-G,-H). Twenty-eight isolates of R. equi from foals with respiratory problems were used in this study. The PCR amplification reaction was performed using specific primers for vap gene family viz. vap-A,-B,-C,-D,-E,-F,-G,-H which yielded the bands of 350, 400, 450, 500, 600, 650 and 700 bp, respectively. All the 28 isolates were positive for vap A,C,D,E,F,G and H and negative for vap B gene which was characteristic of virulent R. equi isolates of equine origin. The multiplex PCR assay for the vap gene family allowed identification and characterization of virulent isolates in a simple one-step reaction. This is a significant advantage in comparison with the conventional diagnostic methodologies of R. equi strains and simultaneously evaluated the presence of the vap A gene which is responsible for virulence in foals.
Keywords | Rhodococcus equi, Equine, vap gene, Virulence- associated protein
Editor | Minakshi Prasad, Head of the Department of Animal Biotechnology, LUVAS, Hisa Lala Lajpat Rai University of Veterinary & Animal Sciences (LUVAS), Hisar, Haryana, 125004, India.
Special Issue | 1(2015) “Biotechnological and molecular approaches for diagnosis, prevention and control of infectious diseases of animals”
Received | March 12, 2015; Revised | April 16, 2015; Accepted | April 17, 2015; Published | April 21, 2015
*Correspondence | Sandip Kumar Khurana, National Research Centre on Equines, Hisar, India; Email: [email protected]
Citation | Chhabra S, Khurana SK, Kapoor PK, Singha H, Singh Y, Khirbat R (2015). Characterization of Rhodococcus equi isolates from foals with respiratory problems using a multiplex PCR for the vap genes. Adv. Anim. Vet. Sci. 3(1s): 28-32.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.1s.28.32
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Chhabra et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
R. equi is Gram positive and facultative intracellular pathogen belonging to order Actinomycetales. These are coccobacillary, aerobic and non-motile organisms formerly known as Corynebacterium equi. Infection causes subacute or chronic abcessating or suppurative bronchopneumonia, ulcerative lymphangitis and enteritis (Meijer and Prescott, 2004) in foals aged 1-4 months (Tkachuksaad and Prescott, 1991; Yager et al., 1991; Khurana et al., 2009; Von Bargen and Haas, 2009; Khurana, 2015a, 2015b; Khurana et al., 2015) months old. R. equi has been reported to infect number of domestic animals including pig, sheep, camel and cattle and has been also recognized as opportunistic pathogen in both immunocompetent and immunocompromised humans and readily cause infection in HIV positive patients (Takai et al., 1995; Mizuno et al., 2005; Napoleao et al., 2005; Khurana, 2014; Khurana et al., 2014). It is commonly isolated from soil, feces and gut from healthy and sick animals. Identification of the virulence plasmids associated with R. equi is of great importance in characterizing pathogenicity of disease in equines (Monego et al., 2009). The virulence is associated with the ability of the bacteria to prevent phagosome-lysosome fusion and replication within macrophages resisting clearance by organism defence (Kanaly et al., 1993; Krewer et al., 2008). Virulence factor is associated with presence of 85-90kb plasmid which encodes various virulence associated antigen (Byrne et al., 2001). According to many researchers virulent and intermittently virulent R. equi is presented with virulence associated antigen Vap A (15-17 kDa) and Vap B (20 kDa) respectively. Gene for the vapA antigen is virtually found in all clinical R. equi isolated from foals (Costa et al., 2006; Takai et al., 1996) and vap B gene on isolates from pigs and humans. Earlier studies identified pathogenicity island within the plasmid which contains seven vap genes and discovery of more vap proteins has taken this number to nine vap genes vap-A, -B, -C, -D, -E, -F, -G, H and I. Thus the presence or absence of the vapA gene can be used as a marker to show presence or absence of the vapA encoding virulence plasmid. Virulence plasmid negative strains of R. equi do not express vapA gene and are therefore incapable of causing disease (Jain et al., 2003). The multiplex PCR assay for the vap gene family allowed identification and characterization of virulent isolates in a simple one-step reaction. This is a significant advantage in comparison with the diagnostic methodologies presented by Halbert et al. 2005 which allowed identification of R. equi strains and simultaneously evaluated the presence of the vapA gene. The PCR technique has the potential to identify virulent R. equi rapidly by amplification of gene sequences that are unique to the virulence plasmids. This approach is useful not only for epidemiological investigations but also for early diagnosis programs. This study was aimed at the evaluation of molecular characteristics of R. equi isolates obtained from foals and adult horses from different geographical locations in India by Multiplex PCR assay that amplifies the vap gene family (vap-A,-B,-C,-D,-E,-F,-G,-H).
Materials and methods
Bacterial Isolates
The study was conducted on 28 bacterial isolates from foals with respiratory problems from Haryana and Rajasthan previously identified as R. equi by biochemical tests.
DNA Extraction
All twenty eight isolates were cultured 5% sheep blood agar and then transferred to nutrient agar and incubated for 24 hours at 37°C.Then the colonies of all 28 isolates were inoculated in BHI broth and incubated at 37°C for 24 hours. The genomic DNA of all isolates from overnight BHI broth culture was isolated by snap chilling. One ml of overnight BHI broth culture of all the isolates was taken in eppendorf tube and centrifuged at 7000 rpm for 5 min. at 4°C and supernatant was discarded. Again one ml of BHI is added to same tube and above step of centrifugation was repeated and supernatant was discarded. 100-200 µl of nuclease free water (NFW) was added to the precipitate in the eppendorf tube and kept in water bath for 5 min. at 95°C and immediately kept in ice for 5 min. The suspension obtained was centrifuged at 7000 rpm for 5 min. at 4°C. The supernatant was stored in fresh eppendorf tube and used as DNA template for Multiplex PCR. The purity of DNA was evaluated by taking the ratio of optical densities (OD) at 260 nm to that of 280 nm, by spectrophotometer (Biorad Smart SpecTM Plus). The samples having OD ratio between 1.7-1.9 were considered having acceptable purity and used in future experiments. For computing the concentration of plasmid DNA, it is known that one OD value at 260 nm corresponds to 50 µg/ml of double stranded DNA. Hence, the concentration of DNA was calculated using the following formula:
DNA concentration (µg DNA/ml) = OD 260 x50 x dilution factor
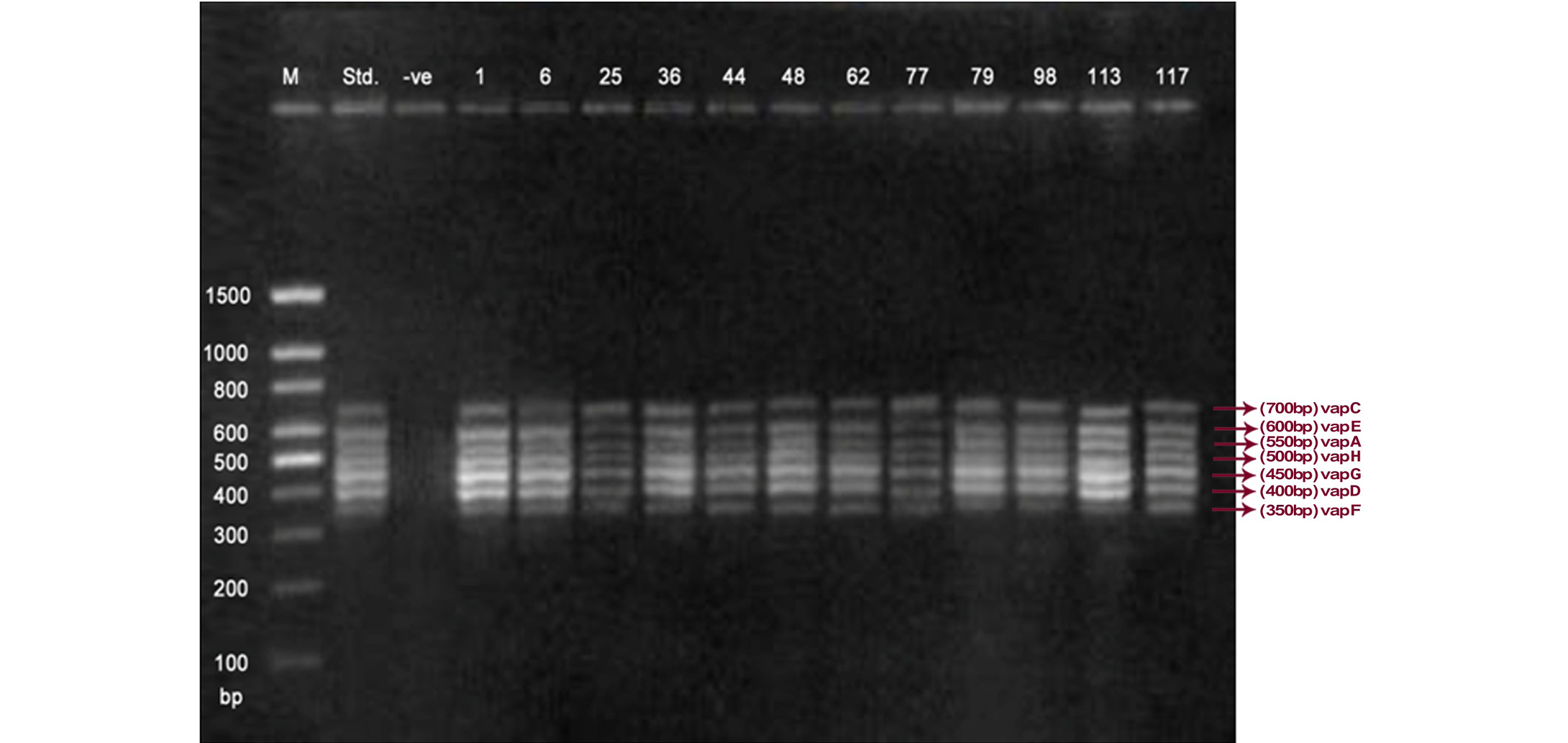
Lane 1: Molecular Ladder; Lane 2: R. equi Standard; Lane 3: Negative control; Other Lane: Respective R. equi isolate number
Table 1: Primers utilized with multiplex PCR for vap gene family amplification
|
Identification |
GeneBank access |
Sequence |
Size (bp) |
|
VAPA-F |
AF116907 |
ACAAGACGGTTTCTAAGGCG |
550 |
|
VAPA-R |
AF116907 |
TTGTGCCAGCTACCAGAGCC |
|
|
VAPB-F |
RER20KDAVA |
GAATTCGAAAGCGCAAAGGT |
650 |
|
VAPB-R |
RER20KDAVA |
TTCCGTGAACATCGTACTGC |
|
|
VAPC-F |
AF116907 |
GGGTCGTCCATCCAAATCGA |
700 |
|
VAPC-R |
AF116907 |
GGTCAGGCCTATCACCCTTG |
|
|
VAPD-F |
AF118814 |
GGTGGTGCGATGTCAGAATG |
400 |
|
VAPD-R |
AF118814 |
TGGAACGTCTTGCCCTTCTT |
|
|
VAPE-F |
AF116907 |
ATATGACGACCGTTCACAAG |
600 |
|
VAPE-R |
AF116907 |
CTCCGATGCCCACCAAACTA |
|
|
VAPF-F |
AF116907 |
AGAATATGCCTGGTATGGGC |
350 |
|
VAPF-R |
AF116907 |
TCGTCGTATAGCTGCTGCAG |
|
|
VAPG-F |
AF116907 |
TCATTGCCACCCTCCGGTTC |
450 |
|
VAPG- R |
AF116907 |
GCGAACGCGGAAACTTCAAT |
|
|
VAPH-F |
AF116907 |
AATTCCTATCAAGGACAGC |
500 |
|
VAPH-R |
AF116907 |
ATACCGATTACGGAGCTCAC |
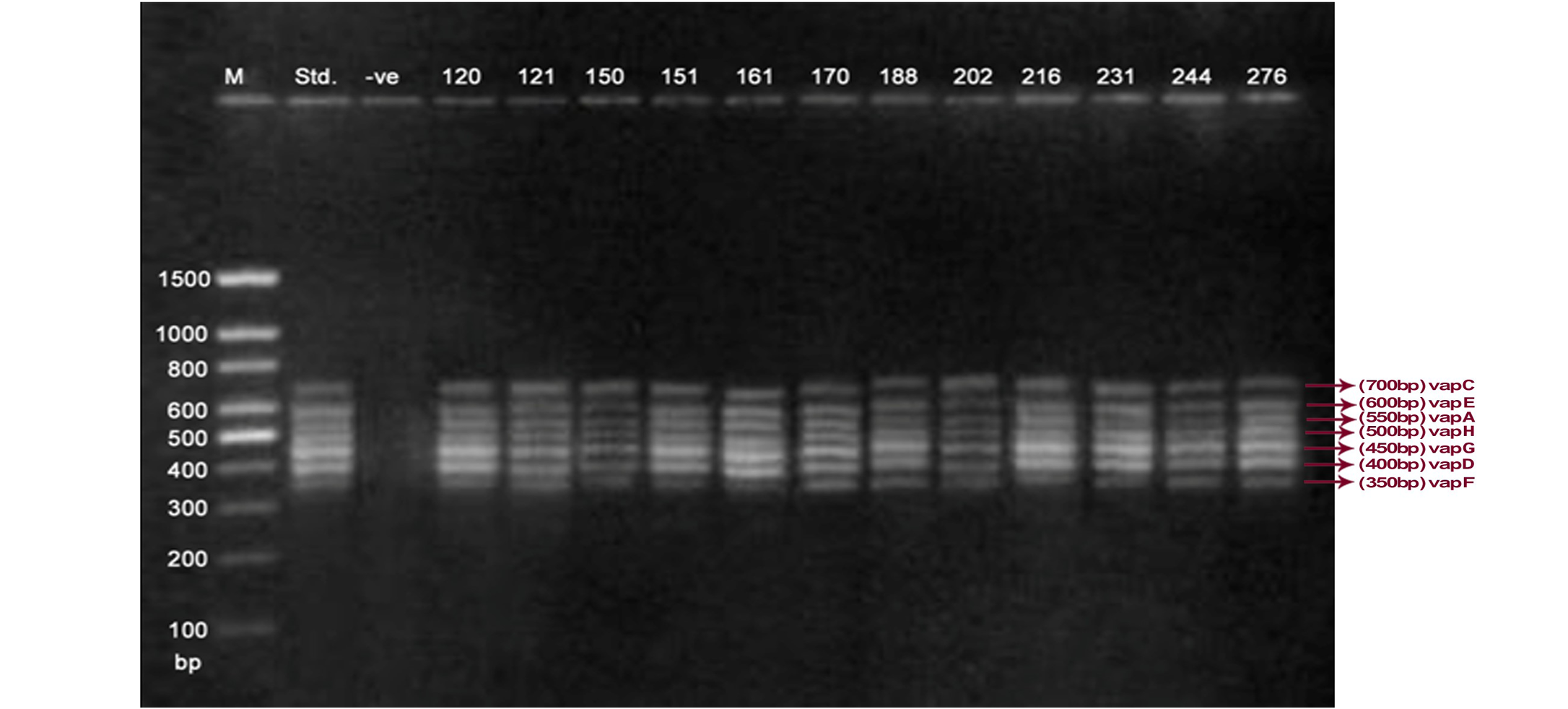
Lane 1: Molecular Ladder; Lane 2: R. equi Standard; Lane 3: Negative control; Other Lane: Respective R. equi isolate number
Multiplex PCR Assay for vap Gene Family
The PCR reaction was carried out as described by Monego et al. (2009) with some modifications. The virulence associated protein (vap) family genes viz. vap-A, -C, -D, -E, -F, -G and –H were amplified using gene specific primers. The details of the primers were mentioned in Table 1. The multiplex PCR cycling conditions consisted of initial denaturation for 5min. at 95°C, denaturation for 45sec. at 95°C, primer annealing for 1 min. at 54°C and extension for 1.5 min. at 72°C following final extension which lasted 10 minutes at 72°C. The samples were subjected to 34 cycles of amplification in thermal cycler (XP gradient cycler Bioer Technology PR, China). For each reaction we used 3.0 μl of DNA preparation in 25 μl reaction mixture containing 10.2 μl NFW, 2.5μl of 10X PCR buffer with 15mM MgCl2 (Fermentas), 1.0μl 10mM dNTP mixture (Fermentas), 0.5μl of 10μM of each forward primers, 0.5μl of 10μM of each reverse primers, Taq DNA Polymerase 0.3 μl (5U/ μl stock)(Fermentas). The amplification products (10μl) were submitted to 1.5% agarose gel electrophoresis at 75 volts for 1 hour 25 minutes.
Results
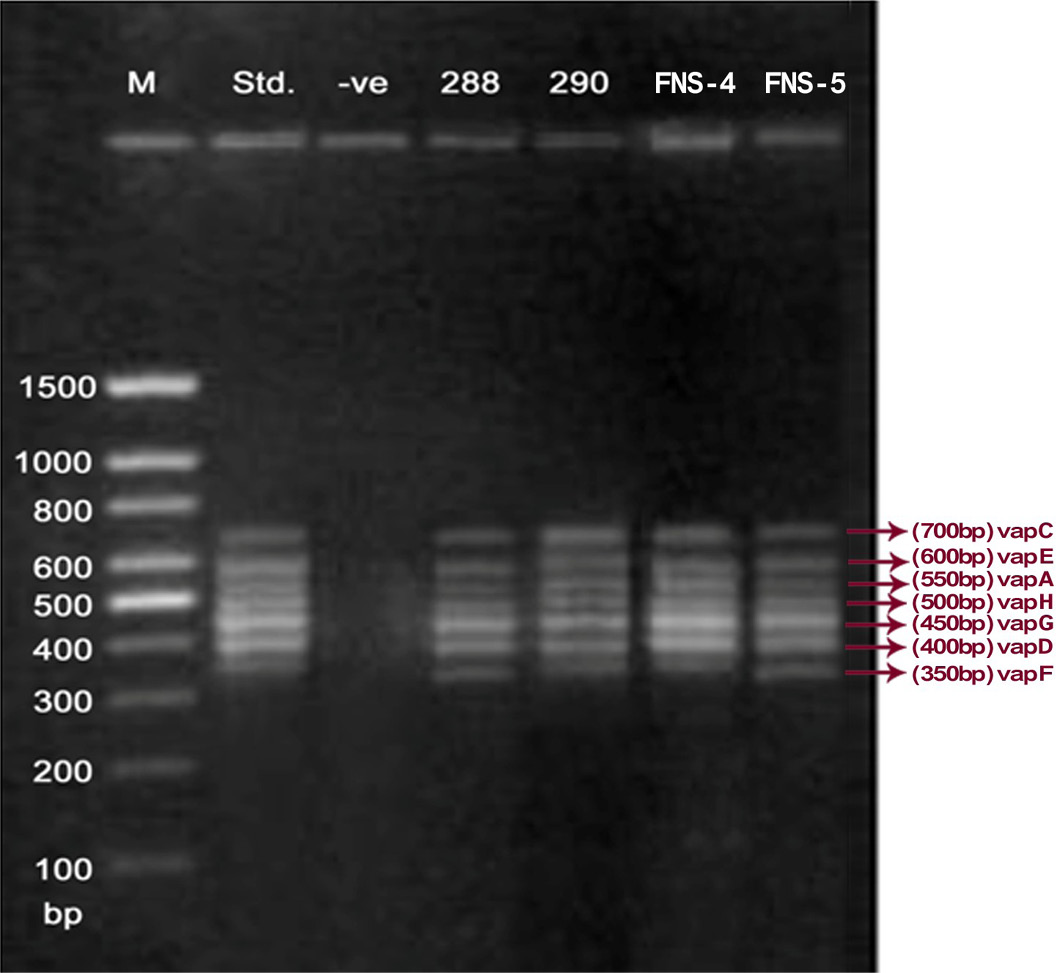
The PCR methodology was applied to analyse 28 isolates. The PCR amplification reaction was performed using the specific primers for vap gene family viz. vap A,-B,-C,-D,-E,-F,-G,-H which yielded the bands of 350, 400, 450, 500, 600, 650 and 700 bp (Figure 1, 2 and 3). The length of DNA fragments amplified by multiplex PCR was in accordance with previous results of Monego et al. (2009). The accuracy of PCR amplification of the vap gene family in our control strains was verified by matching the results with that of standard and negative control used in PCR reaction. All the 28 isolates isolated from foals and adult horses with respiratory problems were positive for vap -A,-C,-D,-E,-F,-G and-H. The amplicon specific for vap-B was absent in all isolates of R. equi.
Lane 1: Molecular Ladder; Lane 2: R. equi Standard; Lane 3: Negative control; Other Lane: Respective R. equi isolate number
Discussion
The virulence associated antigens (Vap-A) and plasmids are used as epidemiological markers for R. equi virulence in foals (Cohen et al., 2005; Takai et al., 1999). Two categories of plasmids: VapA and VapB family are mainly responsible for pathogenesis and host tropism of R. equi (Takai, 1991a, 1991b; Oldfield et al., 2004; Ocampo-Sosa et al., 2007). R. equi isolated from clinical cases express only one of these two plasmids and carries the gene(s) encoded either in vapA or vapB families (Takai et al., 1995). The VapA+ B- type plasmid is associated with the infection in horse (Takai et al., 1995; Ocampo-Sosa et al., 2007). This unique plasmid-determined host-specificity has been found only in Rhodococci, however the precise role and mechanism of the vap antigens remain enigmatic. Monego et al. (2009) identified that none of the 32 vapA-positive isolates showed the presence of vap B gene. The presence of vap-A gene in R. equi clinical isolates and its relationship with lethality of R. equi to susceptible foal was reported by many researchers (Takai et al., 1995; Takai, 1997; Wada et al., 1997). In present study we analyzed 28 R. equi isolates from different geographical regions of India for the presence of vap genes (vap-A,-B,-C,-D,-E,-F,-G,-H) using Multiplex PCR. The molecular profile of the vap gene family found in our study presented vapA, and vapC to vapH genes. The profile all the isolates showed vapA, vapD, and vapG, which in agreement with result data of Jacks et al. (2007), which indicate that these vap genes are the most biologically relevant as they are preferentially induced during infection in the natural host. None of the 28 vap-A positive isolates showed the presence of vap-B gene. The results obtained agreed with that of Takai et al., (1995) reported that R. equi isolates can express either vap-A or vap-B but not both. In our study all isolates were found positive for vap-A gene and negative for vap-B gene, thus providing a vap A+B- profile, which is specific for virulent R. equi isolates of equine origin.
Acknowledgements
The authors are thankful to National Research Centre on Equines, Hisar and Lala Lajpat Rai University of Veterinary and Animal Sciences, Hisar for providing necessary facilities to carry out the research work.
References